- ProductsRequest QuotePackage Products
- DNBSEQ SequencersCycloneSEQ Sequencers
- DNBSEQ Sequencing ReagentsCycloneSEQ Sequencing Reagents
- DNBSEQ Library Prep Kits
- Universal Library Prep Kits
- Modularized Library Prep Kits
- ATOPlex Library Prep Kits
- Single-cell Library Prep Kits
CycloneSEQ Library Prep Kits - mIF Staining Set
- Multiplex Immunofluorescence Reagent
- Bioinformatics Analysis
- Sample Pretreatment Products
- Sample Transfer Platform
- Sample Collection Kit
- Nucleic Acid Extraction Platform
- Nucleic Acid Extraction Kit
- Magnetic Beads
- All Genetic Sequencers
- Sample Preparation Platforms
- Digital Sample Preparation Platform
- Automated Sample Preparation Platform
- Automated Customization Platform
- All Genetic Sequencers
- Integrated Testing Platforms
- Modular Sequencing Workstation
- STOmics Products
- STOmics Product Solution
- Stereo-seq Visualization Reagent
- STOmics Platform
- All Genetic Sequencers
- Ultrasound Products
- Ultrasound System
- All Genetic Sequencers
- BIT Products
- Management system
- Software & Hardware Integrated Machine
- All Genetic Sequencers
- Sample Storage Products
- LT Series
- LN Series
- All Genetic Sequencers
- MGI iLab
- Microneedling Product
- All Products
Recommended for you DNBSEQ-T1+: Sequencing in 24 Hours
DNBSEQ-T1+: Sequencing in 24 Hours T7 Sequencing Kits: Designed for DNA or RNA sequencing on DNBSEQ-T7RS.
T7 Sequencing Kits: Designed for DNA or RNA sequencing on DNBSEQ-T7RS. ATOPlex: Providing panel design and synthetise based on ultra-high plex PCR technology.
ATOPlex: Providing panel design and synthetise based on ultra-high plex PCR technology. FluoXpert Multiplex Immunofluorescence Staining Set
FluoXpert Multiplex Immunofluorescence Staining Set MegaBOLT: Hardware accelerating system for bioinformatics analysis.
MegaBOLT: Hardware accelerating system for bioinformatics analysis. STP-B1000: Precise Identification of Blood Components
STP-B1000: Precise Identification of Blood Components DNBelab-D4: Fully enclosed Library Prep, One Cartridge for All Steps
DNBelab-D4: Fully enclosed Library Prep, One Cartridge for All Steps MGIFLP-L50: New Sequencing Paradigm, All-IN-FLP.
MGIFLP-L50: New Sequencing Paradigm, All-IN-FLP. Stereo-seq Transcriptomics Solution v1.3: Level-up spatial whole transcriptome solution
Stereo-seq Transcriptomics Solution v1.3: Level-up spatial whole transcriptome solution Single-cell RNA Library Preparation Set V3.0: Sensitive detection of genes, 1-4 samples per run to choose from flexibly.
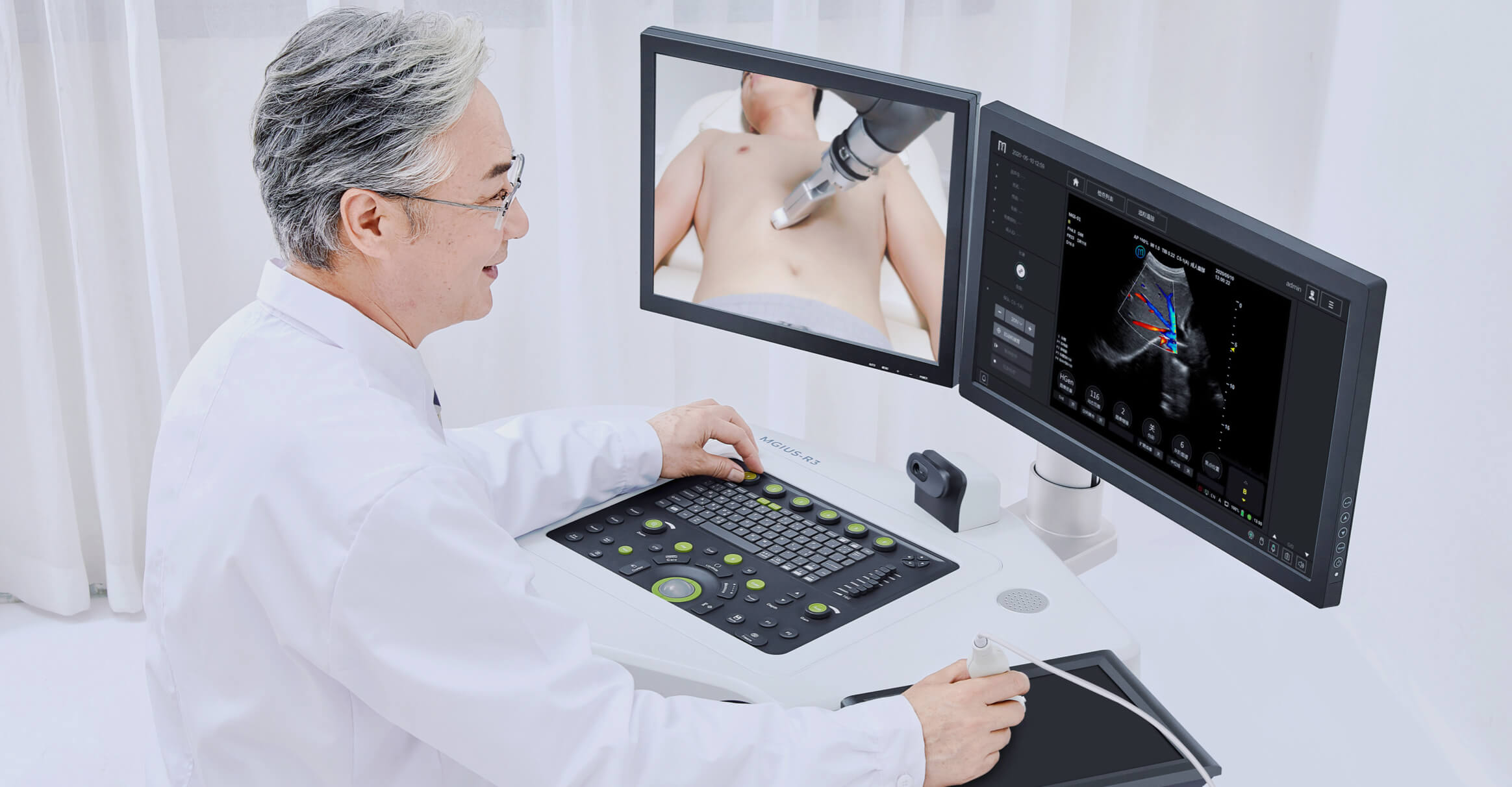
Single-cell RNA Library Preparation Set V3.0: Sensitive detection of genes, 1-4 samples per run to choose from flexibly. R3: Breakthrough with the integration of robotics, teleoperation and ultrasound imaging technology.
R3: Breakthrough with the integration of robotics, teleoperation and ultrasound imaging technology. ZTRON: Promote your Million-scale Genome Project with a Pb-level Sequencing Data Centre.
ZTRON: Promote your Million-scale Genome Project with a Pb-level Sequencing Data Centre. MGICLab-LN55K Pro: Meeting the deep low-temperature storage requirements.
MGICLab-LN55K Pro: Meeting the deep low-temperature storage requirements. Bloomics®: Anytime, Anywhere, Virtually Painless Blood Collection
Bloomics®: Anytime, Anywhere, Virtually Painless Blood Collection MGI's full automation product portfolio
MGI's full automation product portfolio
- Technologies & Applications
- Resources
- About MGI
- 投资者关系
- Products
- Sequencer Products: SEQ ALL
- Genetic Sequencers
- DNBSEQ Sequencers
- T Series
- G Series
- E Series
- CycloneSEQ Sequencers
- WT Series
- Sequencing Reagents
- DNBSEQ Sequencing Reagents
- T Series
- G Series
- E Series
- CycloneSEQ Sequencing Reagents
- WT Series
- Library Prep Kits
- DNBSEQ Library Prep Kits
- Universal Library Prep Kits
- Modularized Library Prep Kits
- ATOPlex Library Prep Kits
- Single-cell Library Prep Kits
- CycloneSEQ Library Prep Kits
- Universal Library Prep Kits
- mIF Staining Set
- mIF Staining Set
- Multiplex Immunofluorescence Reagent
- Bioinformatics Analysis
- Bioinformatics Analysis
- MegaBOLT
- Lab Automation Products
- Sample Pretreatment Products
- Sample Pretreatment Products
- Sample Transfer Platform
- Sample Collection Kit
- Nucleic Acid Extraction Platform
- Nucleic Acid Extraction Kit
- Magnetic Beads
- Sample Preparation Platforms
- Sample Preparation Platforms
- Digital Sample Preparation Platform
- Automated Sample Preparation Platform
- Automated Customization Platform
- Integrated Testing Platforms
- Integrated Testing Platforms
- Modular Sequencing Workstation
- Novel Products
- STOmics Products
- STOmics Products
- STOmics Product Solution
- Stereo-seq Visualization Reagent
- STOmics Platform
- Cell Omics Products
- Single-Cell Products
- DNBelab C Series
- Ultrasound Products
- Ultrasound Products
- Ultrasound System
- BIT Products
- BIT Products
- Management system
- Software & Hardware Integrated Machine
- Sample Storage Products
- Sample Storage Products
- LT Series
- LN Series
- MGI iLab
- MGI iLab
- Microneedling Product
- All Products
- View All Products
- All Products
- View All Products
- Technologies & Applications
- Resources
- About MGI
- Introduction
- Newsroom
- Doing Business With Us
- Creative Club
- Contact Us
- Join Us
- Data Security



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club