MGI's DNA sequencing instruments utilize the state-of-the-art core technology called DNBSEQTM. DNBs (DNA nanoballs) are pumped with by the fluidics system and loaded onto a Patterned Array chip. Sequencing primer is then added and hybridized to the adaptor region of the DNB. The sequencing reaction starts by pumping sequencing reagents containing fluorescently labeled dNTP probes and DNA polymerase. Images are taken after the fluorescently labeled probes on the DNB are excited with lasers. The images are then converted into a digital signal using MGI's propriety software. This information is then used to determine the DNA sequence of the sample.
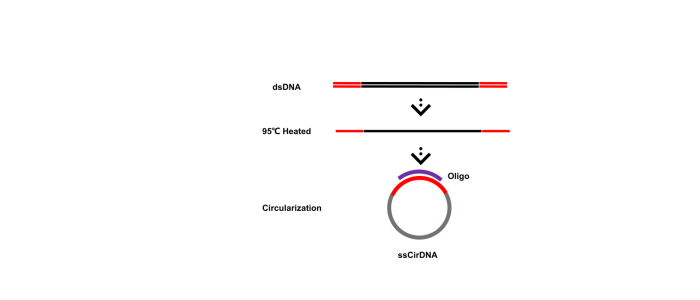
All sequencing technologies relating to DNBs are part of DNBSEQTM. It includes: DNA single strand circularization and DNB preparation technology, Patterned Arrays, DNB loading, cPAS (combinatorial Probe Anchor Synthesis), Pair-End Sequencing technology on DNBs, fluidics and imaging systems, base calling algorithms, etc. cPAS technology has been widely used on various sequencing platforms including BGISEQ-50, BGISEQ-500, DNBSEQ-G50, DNBSEQ-G400, DNBSEQ-T7, etc.
Compared to other existing sequencing platforms, DNBSEQTM sequencing technology combines the advantages from low amplification error rates from DNBs and high density patterned arrays. These advantages dramatically improve sequencing accuracy, and have much lower duplication rates in WGS/WES applications. When combined with the PCR-free library construction method, DNBSEQTM also has higher SNP and indel detection accuracy compared to other platforms. In addition, the index hopping rate in DNBSEQTM platforms is much lower as compared to that of other platforms.

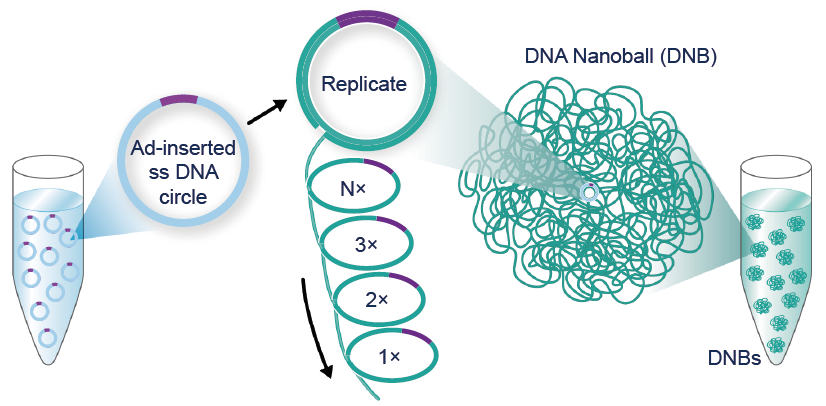
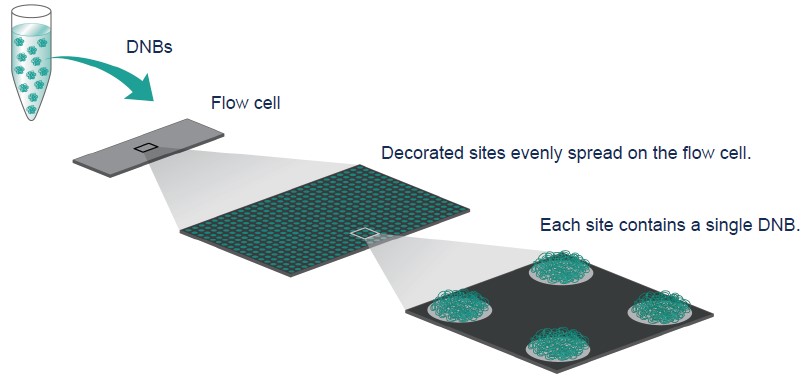
DNBs are optimized so they are the same size as the active sites on the slide surface. This ensures that only a single DNB is loaded onto each active site, which improves effective spot yield.
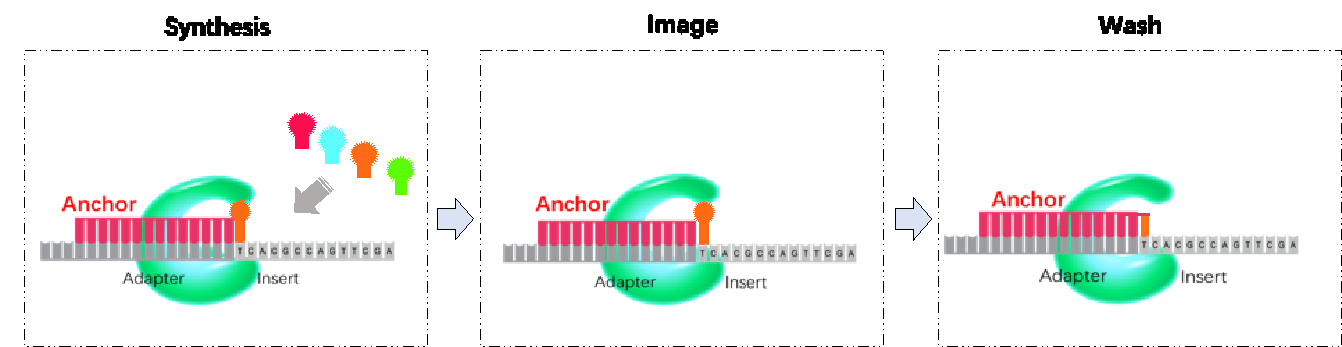
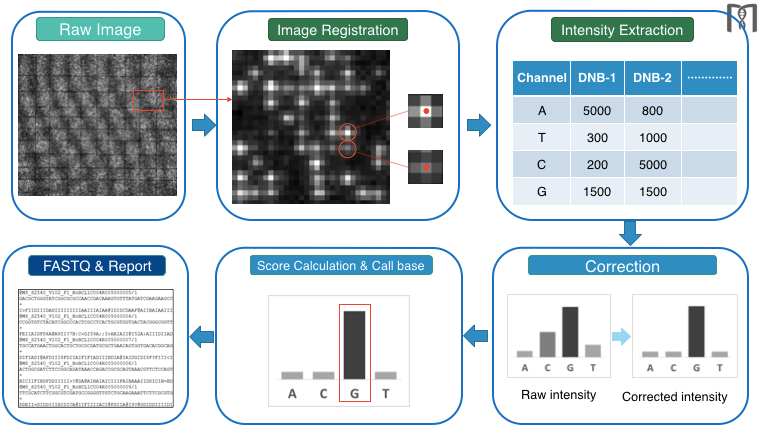
cPAS Technology: After sequencing primers are hybridized to the adapter region of the DNB, a fluorescently labeled dNTP probe is incorporated with a DNA polymerase (Figure 4). Any unbound dNTP probes are then washed away, DNB Flow Cell is imaged (Figure 4: Imaging), fluorescence signal is converted to digital signal, and the base information is determined using MGI's proprietary base-calling software. After the image is taken, regeneration reagent is added to remove the fluorescent dye and prepares the DNBs for the next cycle
The sequencing reaction time has been reduced to less than one minute due to significant improvements in sequencing biochemistry, as well as the identification of a superior sequencing polymerase screened from tens of thousands of mutants.
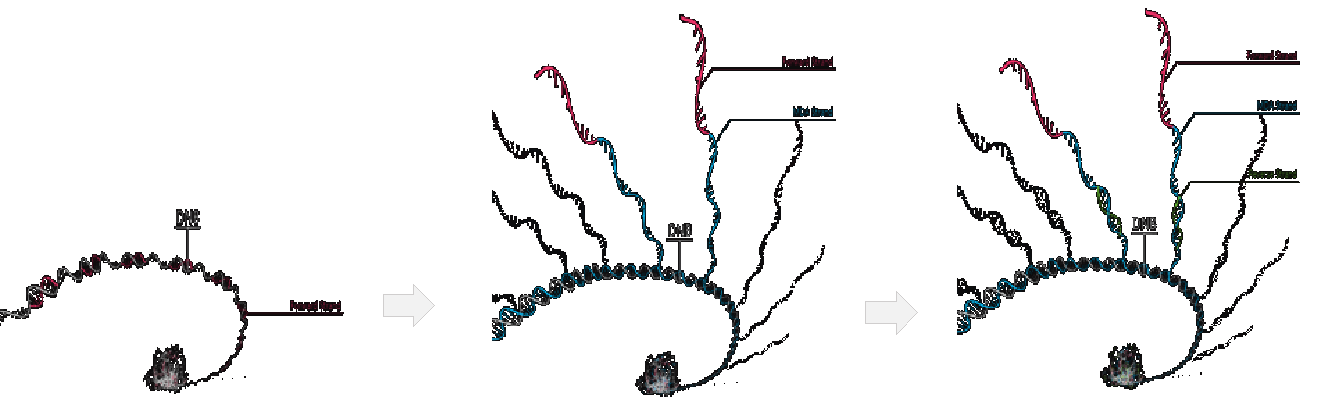
After finishing the 1st strand sequencing, the 2nd strand generation primers and a polymerase with strand displacement activity are added to initiate 2nd strand synthesis. The polymerase will extend the new primer until it reaches the original sequenced strand, at which point it will displace the original sequencing strand to form a new single stranded template. The newly generated 2nd strand is optimized to maximize the length of the strand while ensuring the strand remains attached to the original DNB. After the 2nd strand sequencing primer is hybridized, the same sequencing chemistry is used for 2nd strand sequencing as was used for 1st strand sequencing (Figure 5). The new 2nd strand template has many more copies of insert DNA which yields much stronger signal and increased sequencing accuracy for the 2nd strand.
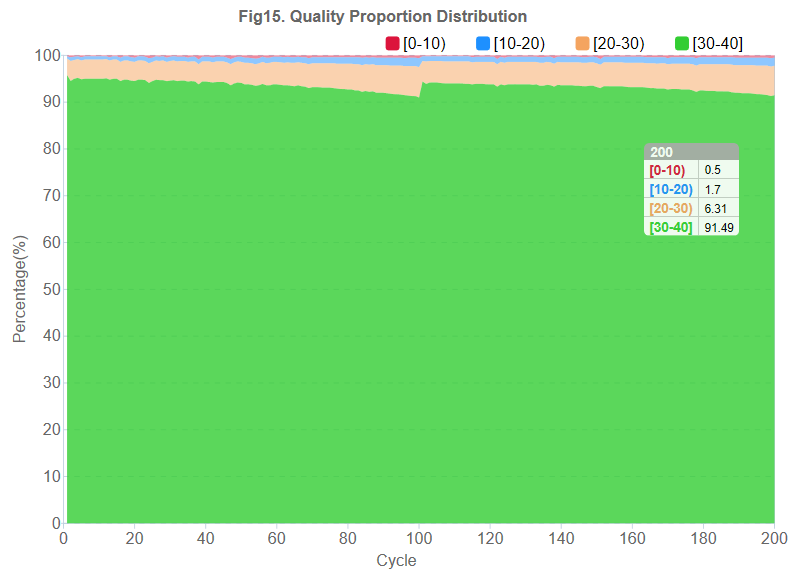
Base calls and base call quality is calculated based on the signal intensities from all channels. The relationship between signal characterization and sequencing error is well established based on known data models. Predicted sequencing errors for unknown samples are calculated based on signal characterization. Quality scores are based on phred-33 standard.
MGI has developed a propriety Sub-pixel Registration algorithm, which enables image intensity extraction at the sub-pixel level, and greatly improves base call accuracy.
Our industry-leading technology has dramatically increased data processing speed and accuracy through integration of a GPU accelerated algorithm, optimization of execution efficiency, and real time image analysis and base calling.



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club







