
T7+: The End of Compromise
As a highly efficient integrated ultra-high-throughput sequencer designed for large-scale sequencing scenarios, powered by MGI's DNBSEQ™ Technology and SM2.0 biochemistry, T7+ can deliver over 14Tb of high-quality data within 24 hours. Its 7-in-1 modular design enables full automation from sample to report, ensuring a streamlined sequencing experience. With an annual capacity of up to 35,000 whole-genome sequencing (WGS), there is no need for compromise between throughput and speed.
Features


Ultrafast Breakthrough: >14 Tb/24 h
|

7-in-1 Intelligent Integrated: Provide a one-stop process, covering DNB making, DNB loading, sequencing, pure water container, waste container, data analysis (including FASTQ files generation and bioinformatic analysis) and data compression software.
|

1-4 Flow Cells: QUAD-Flow Cell Sequencing Independently. PE150 and PE100 at the same time.
|

High-Quality Data Output: Q40>90%
|

Friendly in Use: Ergonomically design, adjustable screen angle, and the reagent cartridge features a modular design, making the preparation process straightforward.
|

Smart System Management: Equipped with comprehensive system sensing, real-time diagnostics, proactive fault detection, and self-healing capabilities.
|

Intelligent Interaction: Omni-smart hub; Smart IP assistant guides workflows.
|

Data Loss Prevention: Resume sequencing at a secure checkpoint following interruption.
|
Why Choose T7+?
- 7 in 1
- A complete workflow from library to report
Minimalist industrial aesthetics; ergonomically designed compartment; and adjustable screen angle. Complemented with an omni-smart hub, making operation as easy as using a smartphone.
*For data generated by the T7+, the compression ratio is expected to be up to 5x vs. gzipped FASTQ.
○ Ultra-High Throughput and Speed
The proprietary TDI camera combined with ultra-high-density flow cells, delivers 100% throughput enhancement while reducing the scanning time by 50%.
- Quad-Flow Cell Sequencing Independently
- More than 14 Tb Data within 24 h*
- Up to 35,000 WGS/Year
*PE150 Read Length


○ Faster, Smarter, and More Functional
Combine DNB making, loading, sequencing, pure water container, waste container, data analysis(including FASTQ files generation and bioinformatic analysis) and data compression into a one-stop process.
○ Q40>90%
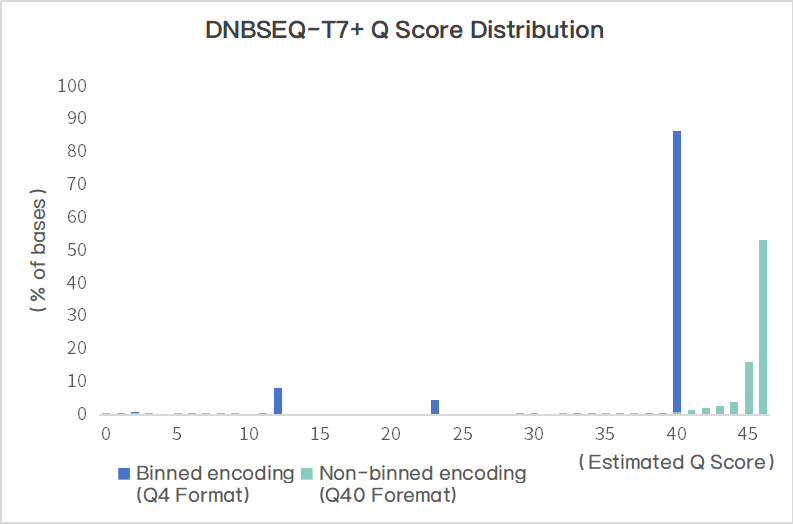
The generated FASTQ files by T7+ adopt a data binning format (Q4 format) for quality scores, which represents the average error rate of the corresponding group. This approach reduces storage space and bandwidth requirements without compromising accuracy or performance. The scores are divided into four groups: N, Q10, Q20, Q30 and above.
*The figure on the right compares the quality scores under non-binned encoding and binned encoding for the same dataset. The data quality is high, with over 90% of bases meeting the Q40 quality threshold.

○ Let Your Sequencer Truly Understand You
○ Lossless Data Compression
Built-in ZARC Data Compression Software, with FASTQ file compression ratio up to 5×.*


○ Empower Multi-Omics
Provide support for multi-application, including WGS, spatio-temporal omics, cell-omics, proteomics, epigenomics, transcriptomics, cancer research and agriculture.
Performance Parameter
|
No. of flow cell per run |
Flow cell type |
Supported read lengths* |
Reads/flow cell** |
Data output per run |
Q30*** |
Run time per run**** |
|
4 |
FCL |
PE75 |
10,000 M~12,000 M |
/ |
>90% |
/ |
|
PE100 |
12,000 M |
2.4 Tb~9.6 Tb |
>90% |
<20 h |
||
|
PE150 |
3.6 Tb~14.4 Tb |
>90% |
<24 h |
|||
|
* The instrument is equipped with PE150, PE100 and PE75 sequencing modes, and the existing reagent kits support PE150, PE100 and PE75 read length sequencing.
** The maximum number of effective reads are based on a specific standard library, and the actual application library will fluctuate depending on the sample type and library construction method. *** The percentage of bases above Q30 is obtained by averaging metrics for runs using standard library. Actual performance is influenced by sample type, library quality and type, insert length among other factors. **** 24-hour run time is from DNB making to cal. files generation (including automated wash). |
||||||
|
Application type |
Requirements |
Recommended read length |
Recommended sample numbers for a single run on T7+* |
|||
|
1*FC |
2*FC |
3*FC |
4*FC |
|||
|
Single cell RNA-Seq |
600 M/sample (C4: 10K cells, 50K reads/cell) |
PE100 |
20 |
40 | 60 | 80 |
|
Cancer large panel |
10 Gb/ sample (5,000X, 1Mb panel) |
PE100/PE150 |
288 | 576 | 864 | 1,152 |
|
Transcriptome |
6 Gb/ sample |
PE150 |
480 | 960 | 1,440 | 1,920 |
|
WGS |
100 Gb/ sample (30× average sequencing depth) |
28 | 56 | 84 | 112 | |
|
WGBS |
110 Gb/ sample (30× average sequencing depth) |
24 | 48 | 72 | 96 | |
|
Stereo-seq |
3,000 M/sample (FFPE) |
PE75 |
3 | 6 | 9 | 12 |
|
1,000 M/sample (FF) |
12 | 24 | 36 | 48 | ||
|
* Sample numbers are calculated considering pooling variation and applications. For reference only. |
||||||
System Parameter
- Power & Dimensions & Net Weight
Power Type: 200 V-240 V~,50/60 Hz
Rated Power: 5,000 VA
Dimensions: 1,370 mm (w)×1,760 mm (h)×848 mm (d)
Net Weight: ~755 kg
- Operating Environment Requirements
Temperature: 15℃~25℃
Relative Humidity: 20%~80%, non-condensing
Atmospheric Pressure: 70 kPa~106 kPa
- Transportation/Storage Environment
Temperature: ~20℃~50℃
Relative Humidity: 15%~85%, non-condensing
Atmospheric Pressure: 70 kPa~106 kPa
Related Products
Related Documents
- Materials
Video



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club





