Recently, the National Health and Medical Commission issued the "Novel Coronavirus Virus Pneumonia Diagnosis and Treatment Plan (Seventh Edition)". The new version of the diagnosis and treatment plan mainly adjusted the following: the epidemic prevention situation prompts vigilance against imported cases abroad; the increase in the transmission route "should pay attention to feces and urine to the environmental contamination caused by aerosol or exposure "; increased description of clinical manifestations of pregnant women and children; increased exclusion criteria for suspected cases ... of which diagnostic criteria are based on two existing laboratory testing methods (RT-PCR nucleic acid detection and viral genes On the basis of sequencing, the new serum-specific coronavirus-specific lgM and lgG antibodies were added as one of the etiological diagnostic criteria.
Regarding the several methods proposed in the guide, what are the differences between them, and what tools and platforms are required in actual applications to be able to successfully carry out the detection? Let us find out together:
Exploring New Coronavirus Detection Methods
Method 1: Real-time fluorescent RT-PCR detection
The characteristics of real-time fluorescent RT-PCR is very fast, and its detection principle is to design specific primers and probes for the specific nucleic acid sequence of the virus to be identified. Most of the kits on the market currently recognize the RdRp gene, that is, the nucleic acid surrounding the virus Copy the gene design probe that expresses that part.
During the replication of real-time fluorescent RT-PCR, this part of the virus sequence was identified by the probe, and then the viral sequence was replicated and amplified by PCR.During the amplification process, dyes were added to the reaction system and signals were captured in real time. Finally, the Ct value is calculated by the software attached to qPCR for analysis of test results.
Theoretically, this method is very sensitive, as long as a virus's nucleic acid is recognized, it can be amplified. The smaller the number of cycles in this process, the more obvious the curve improvement means the higher the load of the virus, so it is a very specific method.
Method 2: viral gene sequencing
If the fluorescent RT-PCR method is like fishing in a pond, then sequencing the genome of a sample is similar to using a large net to catch all the fish in the pond, removing human genes, viral genes and other genes of the microorganisms are all identified.
The metagenomic sequencing method is more comprehensive and sensitive, especially for multiple infections, such as pneumonia patients may also be infected with influenza A, B, mycoplasma, rhinovirus, etc., can also be identified by metagenomic sequencing, but the entire process whose time is longer than the RT-PCR method. In addition, when the viral load is lower than the minimum detection limit of RT-PCR and the background of the human host is low, specific identification can also be achieved by high-depth sequencing.
In addition, the full-length genome of SARS-CoV-2 virus can be obtained based on the multiplex PCR-targeted Panel-based high-throughput sequencing method. This method can complete the detection of multiple targets at the same time as high-throughput sequencing.
In the specific process, the sequencing method is to prepare the library after extracting the nucleic acid, and then enter the sequencer to sequence to obtain the sequence information. After the background data is filtered, it is compared to the database to obtain the complete reads of the virus and finally assembled into a complete Genomic sequence.
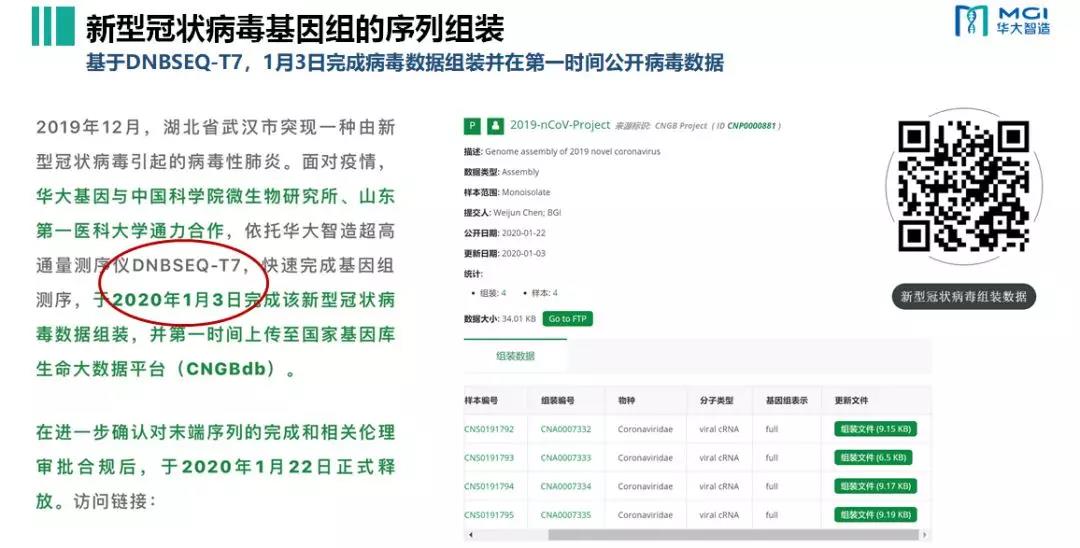
Figure 1.DNBSEQ-T7 to complete the assembly of the new Coronavirus genome on January 3.
Method 3: Serology
The serological test method is simple and convenient to operate, with the fastest results within 15 minutes. However, the sensitivity and specificity of the reagent are relatively limited, and it is suitable for the detection of a large number of suspected cases and asymptomatic infections. Therefore, China currently widely uses Nucleic acid detection.
The three methods of fluorescent RT-PCR, high-throughput sequencing, and serological examination have their own focuses and cannot be replaced. The combination of multiple detection methods can effectively shorten the detection window period and improve the positive detection rate.
Inventory of New Coronavirus detection programs
Focusing on the laboratory detection method of New Coronavirus, let's explore from several levels such as one-stop, automation and high-throughput.
Fluorescent RT-PCR nucleic acid detection is similar to high-throughput sequencing.After receiving the sample; the laboratory must first inactivate the sample, and then use the extraction kit to extract the nucleic acid of the sample. According to the different detection Method, RT-PCR will configure the reaction system, and high-throughput sequencing involves the process of library preparation. Next, they will enter their respective instruments to detect the nucleic acid sequence, and then the system will perform automatic determination and result output.
Among them, the manual operation of the intermediate steps such as sample extraction, system configuration, or database construction can be completed by automated equipment, which is safer and faster.
How to configure devices for different detection processes
So how to configure laboratory equipment specific to different detection processes? Take MGI as an example:
For the RT-PCR detection method, a mgisp-960 can process 96 samples of nucleic acid extraction in one hour (currently upgraded to 80 minutes to process 192 samples), and theoretically can process 500-1000 samples in 8 hours a day; and common fluorescence The pcr is estimated to process 700 samples in an average of 12 hours, and the two devices can be used in a 1: 1 configuration. At the same time, MGI also provides suitable kits, including nucleic acid extraction kits and nucleic acid detection kits configured with a fluorescent PCR system. It can stably achieve a sample processing capacity of at least 500 cases per day.
For the high-throughput sequencing (metagenome) detection method, the limit of the sample size detected by this method is mainly the gene sequencer. Taking the current DHBSEQ-T7, the largest throughput of MGI, as an example, it can process up to 128 samples per day (data (> 80M reads), so 4 MGISP-100 or 2 MGISP-960 can be configured upstream to complete the automated process from extraction to library construction.The adaptation kit can be packaged together.
For high-throughput sequencing (targeted) detection methods, the principle is automated extraction + multiplex PCR targeted sequencing. Unlike the exhaustive metagenomic sequencing, this method uses a special detection kit for new coronavirus to detect samples before sequencing. In the preparation, the genes of the new Coronavirus are taken out and then sequenced. This method combines the advantages of metagenomic sequencing and RT-PCR, while obtaining the full sequence of the viral genome, the sequencing sample size can be increased by 10-20 Times.
On this basis, the internal reference can also be used to relatively quantitatively determine the viral load, which is more suitable for epidemic detection and statistical analysis of virus changes in a large number of samples. At present, MGI has launched a detection kit based on this method (I.e. multiplex PCR sequencing panel).



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













