As a result of the outbreak of COVID-19, this year's 53rd European Congress of Human Genetics (ESHG 2020) was held online. MGI CSO Dr. Rade Drmanac, Dr. Nik Matthews of Institute of Cancer Research, UK, Prof Miguel Esteban at the Chinese Academy of Sciences Guangzhou Institute of Biomedicine and Health, and Prof. Lars Engstrand of Karolinska Institute & Science for Life Laboratory in Sweden shared the latest progress of high-throughput sequencing technologies and valuable experience of utilizing the scientific research ability in the global battle with COVID-19. Among them, Professor Lars Engstrand shared his experience and his team's participation in COVID-19 test and research in Sweden. Professor Engstrand presented the following update and study.
“We started collaboration with MGI at the beginning of April and the lab was established a couple of weeks ago. In the last 4 weeks, we turned our microbiome lab into a high throughput COVID-19 test lab in Stockholm. And as I said, it is urgent to increase the testing and its capacity. We need this not only for diagnosing the individuals that are infected but also for epidemiology and tracking mutations.”
------Prof. Lars Engstrand
Centre for Translational Microbiome Research and the Fight against COVID-19
Faced with the impact of COVID-19, we are urged to improve the capacity of COVID-19 test in Sweden. In late March 2020, we jointly set up a 10,000-person COVID-19 detection laboratory with MGI at the Centre for Translational Microbiome Research (CTMR) of the Karolinska Institute in Sweden, to meet the needs of epidemiological and follow-up studies. CTRM is the National Pandemic Center of Sweden.
In addition to the RT-PCR detection method at CTMR, we specifically set up the high-throughput sequencing platform (DNBSEQ-G400) and ultra-high-throughput sequencing platform (DNBSEQ-T7) for the novel coronavirus sequencing, so as to more comprehensively and promptly map the virus’s spread and detect mutations.
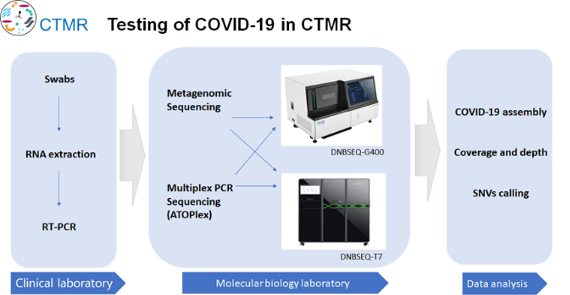
Over 10,000 samples per day, high-throughput and automation are both important
First of all, faced with the need of large-scale sample of RT-PCR detection, relying on high-throughput automated viral nucleic acid extraction equipment can greatly reduce manual operation and improve the speed of large-scale sample detection. At present, CTMR is equipped with 10 MGISP-960, which theoretically meets the needs of detection of 10,000 samples per day.
Second, in the face of the needs of large-scale high flux virus genome sequencing, based on high throughput sequencing platforms DNBSEQ - T7 and DNBSEQ - G400, we respectively used Shotgun sequencing (Metagenomics sequencing) and Multiplex PCR sequencing (ATOPlex) to carry on the further research and analysis on the samples, including COVID-19 assembly, coverage and depth, and SNVs calling.
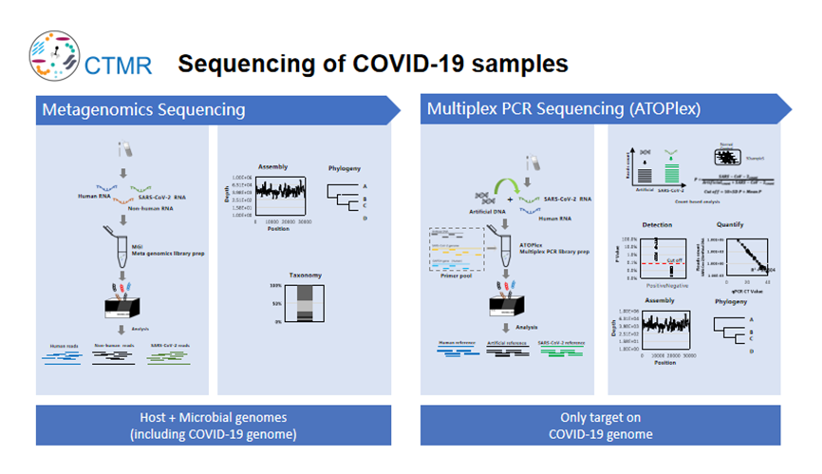
Metagenomics sequencing or Multiplex PCR sequencing (ATOPlex)
Metagenome sequencing can obtain the sequences of the host and microbiome, and can detect unknown pathogens and potential co-infections. Multiplex PCR sequencing is to detect the target virus and obtain the full-length sequence of the target virus based on limited data. At the beginning of the study, the two sequencing methods were analyzed and tested on DNBSEQ-G400 platform. The results of the two methods were consistent that both SNP variation and deletion (18bp) were detected. Both methods can effectively track and monitor virus evolution and mutation.
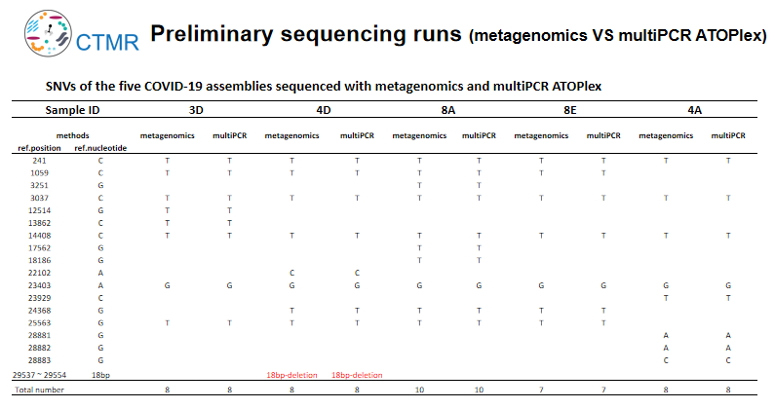
DNBSEQ-T7 or DNBSEQ-G400
In this study, Metagenomic Sequencing was used to sequence the COVID-19 samples based on DNBSEQ -G400 and DNBSEQ -T7 sequencing platform. The sequencing results showed that although the sequencing depth was different, the sequencing results of the same sample in the two platforms were basically the same. The results illustrated that there is good consistency between DNBSEQ sequencing platforms. If large-scale detection is required, the ultra-high throughput sequencing platform DNBSEQ-T7 can be directly selected to complete 128 samples of the Metagenomics sequencing (PE100, 100M reads/ samples) or the 3,840 samples of multiplex PCR amplification (ATOPlex, 5M reads/ samples) in one single run. If the sample size is limited, DNBSEQ-G400 can be chosen for its lower cost per run. In summary, the choice of sequencing platform depends on the sample size per day.
Multi-omics research on COVID-19
Scientists all around the world are working actively on testing and research on COVID-19. Multi-omics research on COVID-19 include the research on Host Genome, Metagenomics, Virus Genome, Single cell, and Proteomics, which will be the focus of CTMR for the present and foreseeable future. At the same time, we are validating the potential of carry out large-scale COVID-19 test and screening in Sweden. I am confident that based on high-throughput sequencing, multi-omics research will play an indispensable role in the global fight against COVID-19.
More information: https://en.mgitech.cn/resource/webinars_info/33/



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













