As today's most common gene-editing system, CRISPR-Cas9 has great potential for applications in the treatment of cancer and genetic disease treatment, as well as animal and plant gene research. Although the specific cleavage of DNA has the unique advantage of being highly efficient for high-targeted modification, multi-target editing, and editing of RNA, CRISPR/Cas has obvious disadvantages: if there is no PAM (Protospacer Adjacent Motif) near the target gene, editing cannot be achieved, and there is also the problem of off-target effects.
Off-target effects are the main limiting factor affecting the widespread application of gene-editing technology. How to correctly assess and detect off-target effects and propose corresponding strategies to reduce off-target effects is one of the most important research directions in the current field of gene editing. Studies have shown that whole-genome sequencing is one of the simplest and most effective methods to detect off-target effects. In 2015, a research team of the Korean Institute of Life Sciences and Technology published an article in Nature Methods "Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells," which emphasized the key role of whole-genome sequencing in identifying the off-target effects of CRISPR-Cas technology and proposed the concept of Digenome-seq for the first time.

Figure 1. Article published in Nature Methods
Recently, the same scientific research team in South Korea published the latest results of their research into gene editing in Nature Biotechnology, using the DNBSEQ-T7 platform* (MGI) to verify CRISPR-Cas12f1 off-target effects based on Digenome-seq sequencing. The researchers developed an effective and specific genome-editing tool, CRISPR-Cas12f1, by remodeling the gRNA. DNBSEQ-T7 whole-genome sequencing data confirmed that the system has high specificity and significantly decreased off-target sites. When delivered using AAV vectors, the CRISPR-Cas12f1 system enables multiplexed and efficient genome editing.

Figure 2. Article published in Nature Biotechnology![]()
CRISPR-Cas12f1 system upgrade
One of the keys to the off-target effects of CRISPR/Cas lies in the design of the gRNA. If there is too much non-specific binding between the gRNA and non-target regions, it will lead to higher off-target efficiency. Therefore, the design and remodeling of gRNAs can help increase the capabilities of CRISPR tools in terms of simplicity, multiplexing, and specificity. To optimize the Cas12f1 gRNA, the research team identified five potential modification sites (MSs) in the tracrRNA and crRNA sequences for remodeling using their background knowledge of gRNA modification, and finally obtained a powerful and very compact CRISPR- Cas12f1 system. The data shows that the engineered gRNAs can significantly increase the indel frequency of most target gene loci, and the average efficiency of the induction by one gRNA was even increased by 867-fold.
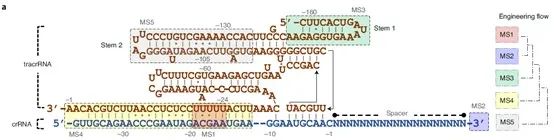
Figure 3. Overall gRNA remodeling strategy, source: Nature Biotechnology
In addition to the compactness of Cas12f1, this system has an additional advantage for gene therapy: it can induce dsDNA cleavage outside the protospacer sequence. This property means that even after the first round of mediated indel mutations, the protospacer sequence is likely to remain unchanged and the dsDNA cleavage process can continue.
Specificity of CRISPR-Cas12f1 gene-editing
Considering the persistent activity of Cas12f1 in cells, it is particularly important to detect the specificity of the CRISPR-Cas12f1 system. The research team used Cas-OFFinder (a potential off-target site prediction algorithm) to identify potential off-target sites and found that Cas12f1 had higher on-target efficiency at target sites and intergenic regions.
Digenome-seq is a whole-genome sequencing technology based on in vitro nuclease digestion and can be used to profile off-target effects of whole-genome nuclease in cells. Digenome-seq has proven to be a robust, sensitive, unbiased, and cost-effective method for analyzing genome-wide off-target effects in gene editing.
The research team performed off-target detection based on Digenome-seq to further determine the specificity of CRISPR-Cas12f1 and performed deep whole-genome sequencing on purified genomic DNA using DNBSEQ-T7*. Scoring DNA cleavage at every nucleotide position across the genome and identifying potential off-target sites. The results showed that not only were fewer off-target sites identified for the Cas12f1 system, but the off-target/on-target ratio was also lower.
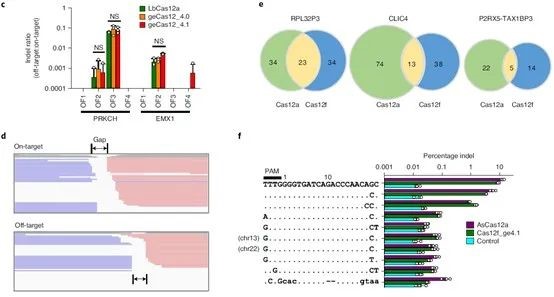
Figure 4. CRISPR-Cas12f1 specific detection, source: Nature Biotechnology
The potential of CRISPR-Cas12f1 for gene therapy applications
The researchers explored the therapeutic efficacy of AAV and CRISPR-Cas12f1 and found a pair of potent sgRNAs when testing on both sides of the c.2991+1655A>G mutation site. An AAV vector carrying the Cas12f1 system was constructed using these sgRNAs and the efficiency of Cas12f1-induced deletion in HEK293T cells was evaluated and compared. It was found that the deletion level induced by the Cas12f system was higher and the deletion rate of the Cas12f1 gene was about 46% higher than that of the control group. This suggested that Cas12f1 may provide a versatile and efficient genome editing system for gene therapy.
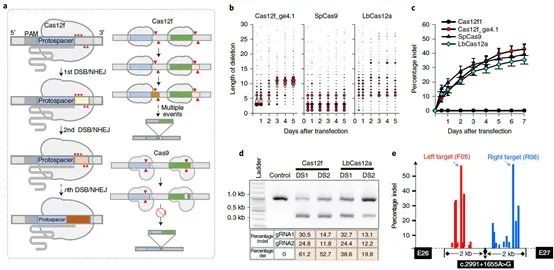
Figure 5. CRISPR-Cas12f1 can integrate with AAV, source: Nature Biotechnology
In this latest study, MGI's DNBSEQ-T7* has successfully completed the task of sequencing the whole genome. The researchers reached a key conclusion based on the sequencing data: the CRISPR-Cas12f1 system has lower off-target effects, indicating that the DNBSEQ-T7 sequencer* performs consistently and can produce high-quality, whole-genome sequencing data.
Accurate and error-free whole-genome sequencing data is the key to forming correct conclusions and high-quality sequencing data mainly relies on library construction as well as robust and efficient sequencers. The ultra-high-throughput gene sequencer DNBSEQ-T7* launched by MGI in 2018 supports whole-genome sequencing, with a daily data output of up to 6 Tb and can sequence the complete genomes of up to 60 humans in one day. The above results show that the DNBSEQ-T7* sequencing platform can provide high-quality whole-genome sequencing data for testing the results and specificity of gene editing, as well as identifying off-target effects.
In addition to DNBSEQ-T7*, other sequencing platforms made by MGI have also had important applications in the development of CRISPR gene editing. In 2020, a team at the Neuroscience Research Institute, Peking University published an article in Science Advances, using the DNBSEQ-G50* to deep sequence a library of gene editing targets and potential off-target sites, in order to verify the specificity of the newly developed gene-editing system.
In the same year, a team at the BGI Group, Shenzhen, published the DocMF platform based on MGI's DNBSEQ sequencing platform* in Science Advances. This platform is the only high-throughput platform in the world that is compatible with identifying protein-DNA binding or cleavage target sites, also has the potential to identify CRISPR system off-target sites and transcription factor binding sites.
References:
[1] Kim, D.Y., et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nature Biotechnology 40, 94–102 (2022).
[2] Xianji.com: Gene editing “wherever you say,” four off-target detection methods. https://www.xianjichina.com/news/details_186013.html



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













