As technology evolves with each passing day, the iteration of tool updates based on technology is even more dazzling. In the life sciences industry, the technology developed from the initial Sanger sequencing to NGS sequencing, from high-throughput sequencing to single-molecule sequencing. Some people mark them as the first and second generation, while the others say third and fourth, or even "2.5 generation". Then, how are these so-called intergenerational relationships divided, and what are the criteria for division? Does it mean each tech has its own strengths, or is there really an "intergenerational relationship"?
What is the intergenerational division standard?
Currently, the industry-recognized unified division standards are only "first generation" and "next generation", which are used to distinguish between Sanger sequencing and non-Sanger sequencing. These two types of technologies have large differences in sequencing principle and sequencing throughput, but they also have similarities. For example, whether it is Sanger dideoxy nucleotide sequencing, or edge-synthesis sequencing in high-throughput sequencing, or sequencing-based sequencing, the principles all rely on nucleotide polymerization.
In the next generation of sequencing technology, how to distinguish between "second generation" and "third generations"?
The "third generation sequencing" formulation emerged in 2008-2009, when it mainly referred to new sequencing technologies that were different from large-scale parallel sequencing. Some scholars believe that single-molecule sequencing, real-time sequencing and core methods are different from prior art methods, which may be the defining characteristics of next-generation sequencing technologies. Currently, second generation sequencing usually refers to high-throughput, large-scale and parallel sequencing, while third generation sequencing usually refers to single-molecule sequencing without DNA amplification.
However, some scholars have pointed out that the current intergenerational division of sequencing technology may be for the sake of commercial reasons, because people tend to believe that technological intergenerational upgrading represents the evolution of technology. In fact, single-molecule sequencing technology was first published as a conceptual paper in 2003 (high-throughput sequencing technology has entered on the market since 2005). In 2008, Helicos BioSciences launched its first single-molecule sequencer, followed by Pacific BioSciences and Oxford Nanopore to launch their own commercial sequencers. However, perhaps because single-molecule sequencing requires more on the technical system, the development of this technology was far less rapid than people expected, and it has not yet reached the market scale of high-level sequencing technology today. During this period, Helicos BioScience went bankrupt in 2012, although its technology was fully consistent with the current definition of three generations of sequencing technology.
Conclusion
Sequencing technology is still in rapid development. The intergenerational division of sequencing technology at this stage lacks uniform industry standards to a large extent. Undoubtedly, both Sanger sequencing and non-Sanger sequencing (NGS, next-generation sequencing or high-throughput sequencing) have led the revolution in genomic technology, driving advances in genomics. Sanger sequencing has made major contributions to the Human Genome Project (HGP) and is still a routine technique in many biological and medical laboratories. Non-Sanger sequencing is the mainstream technology of current genome research and application, which directly clears the economic obstacles for the wide application of genome sequencing, making it not only better serve scientific research, but also becoming important in precision medicine and other application fields. In addition, single-molecule technology is one of the important directions in the development of sequencing technology. Under the constant attempts and efforts of the industry peers, it has finally begun to emerge, but it will take time to mature and improve. These sequencing technologies have their own characteristics, and also have suitable application scopes and application scenarios.
Bonus: a picture to understand the progress of sequencing technology
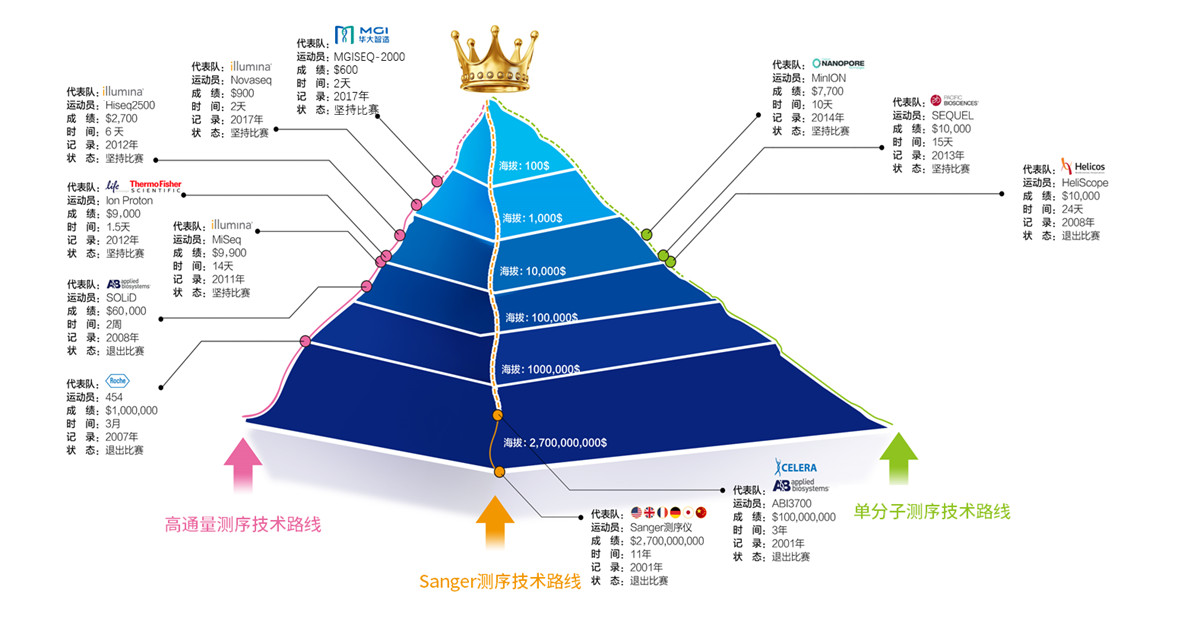
Reference:
【1】Ido Braslavsky, Benedict Hebert, Emil Kartalov, Stephen R. Quake. Sequence information can be obtained from single DNA molecules.[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(7):3960-3964.
【2】Wangwei,2019,http://blog.sina.com.cn/s/blog_bcb043950102zbn2.html
【3】Goodwin S, Mcpherson J D, Mccombie W R. Coming of age: ten years of next-generation sequencing technologies[J]. Nature Reviews Genetics, 2016, 17(6):333-351.



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL














 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













