RNA is an inevitable object of study to use in investigating biological molecular mechanisms. Intracellular RNA contains a wealth of information, including sequence, abundance, structure and so on. Due to the temporal and spatial specificity of RNA expression, sample acquisition is difficult, and accurate analysis of RNA in samples after sampling is particularly important.
Current major RNA research strategies include transcriptome sequencing (mRNA), long-chain non-coding RNA sequencing (lncRNA), small RNA sequencing (small RNA) and circular RNA sequencing (circRNA). MGI has released four RNA library preparation kits based on DNBSEQ technology and adapted to the MGI sequencing platform (Table 1) for the systematic study of RNA.

In the previous issue, we have introduced the MGIEasy RNA Library Preparation Set. This issue focuses on the MGIEasy RNA Directional Library Prep Set. Compared to conventional RNA sequencing library, RNA directional sequencing library saves the orientation information of the RNA strand into the library when constructing the sequencing library. The sequencing data analysis can identify whether the transcript is given from the sense or antisense DNA strand.
Identifying strand origin increases the percentage of reads that align to the transcriptome and provides more accurate information for studies in regulation of gene expression and gene functional analysis. Maintaining strand orientation also allows identification of antisense expression and novel genes. RNA sequencing based on directional sequencing library has been an important means to more accurately study gene structure and regulation of gene expression.
MGIEasy RNA Directional Library Prep Set provides an efficient package for generating RNA directional libraries suitable for MGI high-throughput sequencing platforms. So, what exactly is the performance of the MGIEasy RNA Directional Library Prep Set. Let us explain the features one by one.


1. The input amount of total RNA can be as low as 10ng
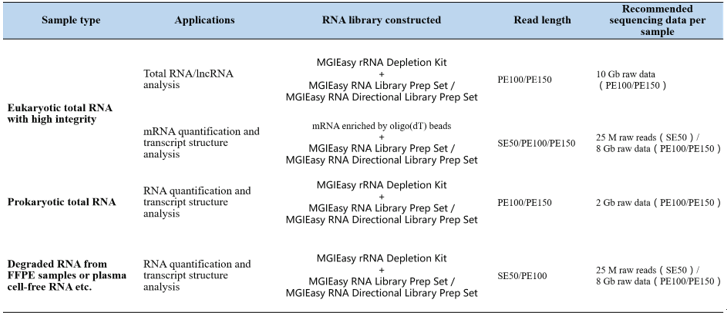
The UHRR (Universal Human Reference RNA, Agilent, 740000) was used to prepare RNA directional library at different total RNA input amount of 10 ng, 50 ng, 200 ng and 1 μg. Libraries were prepared from the mRNA enriched by oligo(dT) beads using the MGIEasy RNA Directional Library Prep Set and sequenced on MGISEQ-2000 at PE100 read-length.
The results showed that the genome mapping and gene mapping rate were >90% and >60% respectively and the numbers of genes or transcripts detected were similar and in high proportions across all input amounts. There was no significant difference between the different input amounts. Even when the input amount is as low as 10ng, the MGIEasy RNA Directional Library Prep Set exhibits excellent quality sequencing data and transcript annotation. Therefore, this is a very helpful solution, which can solve precious samples with less or even only 10 ng total RNA to obtain high quality data.
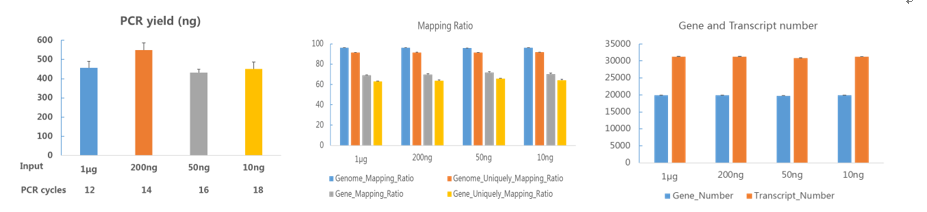

Fig.1 The PCR yield in differentinput amount of total RNA
Fig. 2 The mapping ratio in different input amount
2. Quantitative and accurate gene expression, high repeatability
As shown in Fig. 4 and Fig. 5, the gene expression level (FPKM) of libraries prepared from MGIEasy RNA Directional Library Prep Set and MGIEasy RNA Library Prep Set were consistent and the concordance was above 0.99. The MGIEasy RNA Directional Library Prep Set and qPCR had a consensus of 0.86 in terms of gene quantification accuracy. These results indicate that the MGIEasy RNA Directional Library Prep Set can accurately quantify RNA expression and be applied to analysis of gene expression.

Fig.4 Concordance of MGIEasy RNA Directional Library Prep Set and qPCR quantitative
3. RNA structure analysis is more direct and reliable
In addition to RNA quantitative analysis, MGIEasy RNA Directional Library Prep Set also has advantages in structural analysis. As shown in Fig. 6, the library constructed from MGIEasy RNA Directional Library Prep Set has high strand specificity with a sense strand rate above 99%.
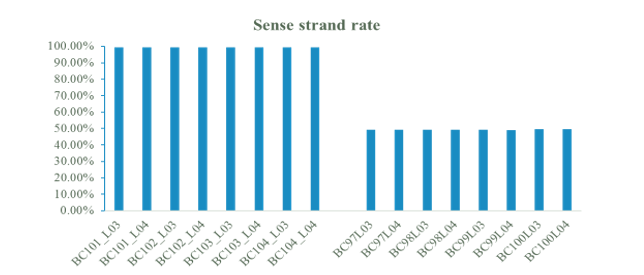

Fig.6 Sense strand rate of MGIEasy RNA Directional Library Prep Set and MGIEasy RNA Library Prep Set
Since the MGIEasy RNA Directional Library Prep Set is able to identify RNA stranded information, it can more accurately locate the junction site under the same read depth (Fig. 7). This is helpful for transcript structure detection and mutation analysis.
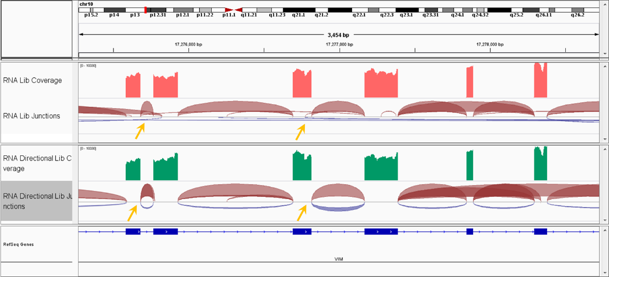

Fig. 7 Transcript structure analysis of MGIEasy RNA Directional Library Prep Set and MGIEasy RNA Library Prep Set
4. High uniformity
Uniformity of coverage across the entire transcript is important for assembly of transcripts and analysis of gene structure. A comparison was performed between the MGIEasy RNA Directional Library Prep Set sequenced on MGI platform and comparable RNA kits sequenced on I platform. The results in Fig.8 demonstrate that the libraries constructed using the MGIEasy RNA Directional Library Prep Set have high uniformity of coverage from transcripts 5' to 3' ends and have superior 3’end coverage over others.
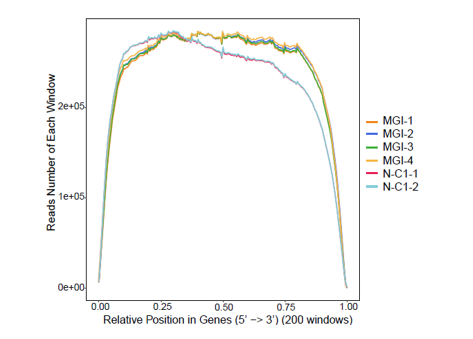

Fig. 8 The reads randomness of different platforms and kits kit
The dual advantages of MGIEasy RNA Directional Library Prep Set in gene structural analysis and quantitative analysis of gene expression make gene detection more accurate.
Of course, we can't just look at the comparison data on the MGISEQ platform. In order to analyze whether there are significant differences between different sequencing platforms, the concordance analysis between the MGIEasy RNA Directional Library Prep Set sequenced on MGI platform and comparable RNA kits sequenced on I platform was performed, and the results show that the gene expression level (FPKM) of MGI libraries and other libraries from I platform were consistent and the correlation was above 0.95 (Fig. 9). Overall, there is comparability between the MGIEasy RNA Directional Library Prep Set on MGI platform and comparable RNA kits on I platform.
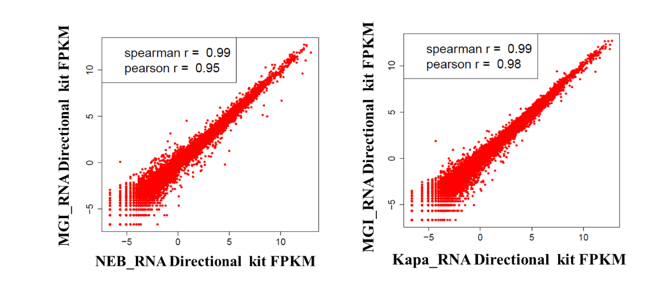

Fig. 9 Concordance of different platforms and kits
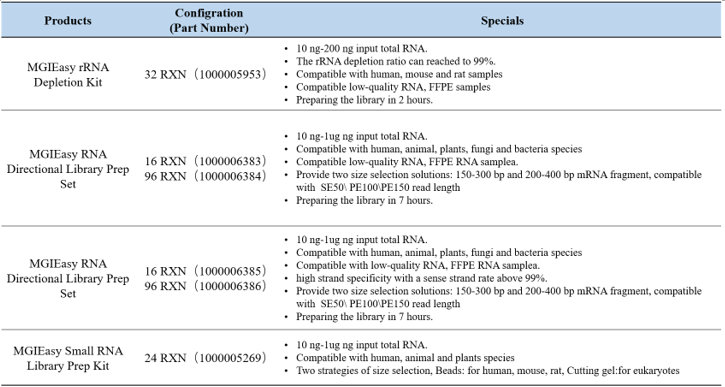

Here's a quick summary of MGI RNA sequencing library preparation products for your choice, so that you can get valuable data from precious samples more easily.
The rRNA Depletion Kit is a highly efficient RNA enrichment kit that can efficiently remove rRNA from total RNA and keep mRNA and non-coding RNA. Another method of RNA enrichment is to capture mRNA with polyA tail and part of lncRNA through OligodT magnetic beads. This method has many mature commercial kits. With the total RNA sample enriched by any of the above enrichment methods, the MGIEasy RNA Directional Library Prep Set or the MGIEasy RNA Library Prep Set can be used for library construction.
Finally, the Demo data of the MGIEasy RNA Directional Library Prep Set is available for test. Please click here:
ftp://ftp.cngb.org/pub/CNSA/CNP0000288/CNS0040524/CNX0048125/CNR0066420/
ftp://ftp.cngb.org/pub/CNSA/CNP0000288/CNS0040524/CNX0048127/CNR0066422/
References:
[1] Salmena, L., Poliseno, L., Tay, Y., Kats, L., & Pandolfi, P. P.(2011). A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language Cell, 146(3), 353–358.
[2] Kallen A N, Zhou X B, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs[J]. Molecular cell, 2013, 52(1): 101-112.
[3] Jin X, Feng C, Xiang Z, et al. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis[J]. Oncotarget, 2016, 7(41): 66455



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













