At the Advances in Genome Biology and Technology (AGBT) General Meeting 2024, MGI introduced the Standard MPS 2.0 (SM 2.0) update to their line of DNBSEQ™ platforms to deliver improved sequencing quality as measured by the percentage of Q40 bases. It not only effectively improved the sequencing data output and parameters such as Q30, but also bring the sequencing quality of DNBSEQ platform into the era of Q40 (>85%).
The debut of StandardMPS 2.0 sequencing reagent heralds a new era of sequencing quality reaching up to Q40 within the genetic sequencing industry, delivering a fresh and enhanced product experience to customers worldwide. Here, we present a comprehensive overview encompassing MGI’s initial findings along with results from two customers utilizing SM 2.0 on the DNBSEQ sequencing platform. The data indicated that StandardMPS 2.0 facilitates DNBSEQ sequencing platform reaching Q40 (>85%). Moreover, whole genome sequencing (WGS), single-cell, microbiological, and other studies can also benefit from the improvement of sequencing data
quality.
How does StandardMPS 2.0 realize Q40?
- DNA Polymerase and MDA Enzyme Optimization
Optimization is achieved by utilizing better DNA
polymerase with a higher polymerization efficiency and MDA (Multiple Displacement Amplification) enzyme with a better second-strand displacement function.
- dNTP Optimization
The base-calling accuracy has been significantly improved through base crosstalk reduction based on the optimization of fluorescent dyes on dNTP.
- Algorithm Optimization
The high-fidelity enzyme and PCR-Free library preparation methods are used to reduce the bias introduced by errors in upstream experiments.With more accurate model training, the precision of data interpretation has been improved, ensuring that the sequencing results more closely match empirical base-calling accuracies.
- Compatible with various DNBSEQ platforms
StandardMPS 2.0 is compatible with various genetic
sequencers (DNBSEQ-G99, DNBSEQ-G400, DNBSEQ-T7, etc.) to meet the demands of customers
in different scenarios.
What if sequencing quality reaches Q40?
Q40 is defined as 99.99% accuracy in base calling1, meaning the base-calling accuracy has been improved from Q30 (99.9%) to a new level. Sequencing quality reaching Q40 will provide more possibilities for scientific research and clinical applications. For example, more accurate detection of low-abundance bases will facilitate the identification of low-frequency mutations in oncology research2, low-frequency drug-resistant mutations in infectious disease research3, and novel
biomarkers such as ctDNA in liquid biopsy4,5. Meanwhile, sequencing quality reaching Q40 is also helpful for the identification of variants such as SNP and InDel in WGS research6, and it can improve the identification accuracy of bacteria and other microorganisms7.

Data exhibition: Effectively promote the research of WGS, single-cell, and microbiology
Data source: MGI
Sequencing platform: DNBSEQ-G99, DNBSEQ-G400, DNBSEQ-T7
Applications: WGS, oncology
Significantly improved sequencing quality
SM 2.0 combined with different DNBSEQ platforms (DNBSEQ-G99, DNBSEQ-G400 and DNBSEQ-T7) can realize a >85% Q40 with improved total reads, Q30, Q40 and other sequencing parameters than SM (Table 1, Figure 1).
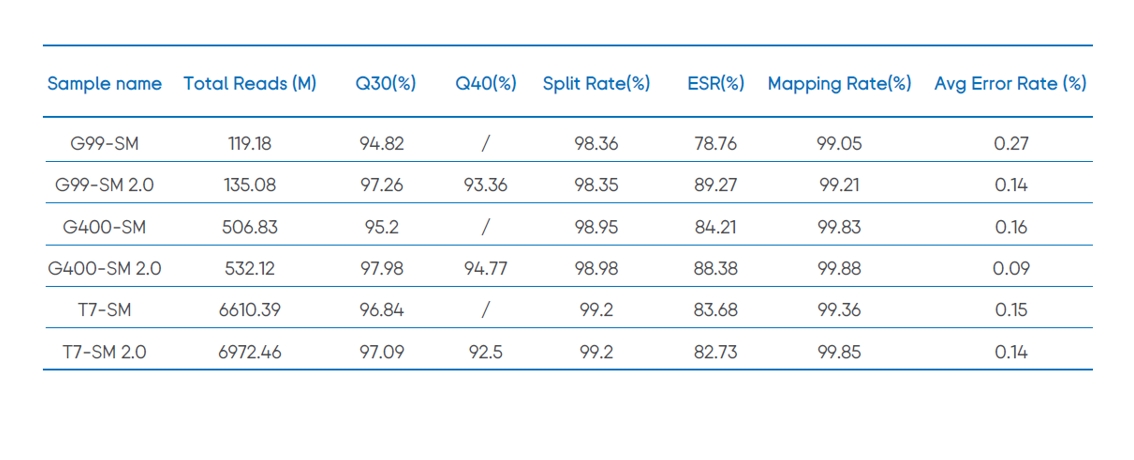
Table 1. SM 2.0 can significantly improve the sequencing quality of DNBSEQ platform.
Note: ESR (%) means Effective Spot Rate, ESR = (Total Reads / DNB Number) ×100%; The libraries above are all E. coli WGS libraries.
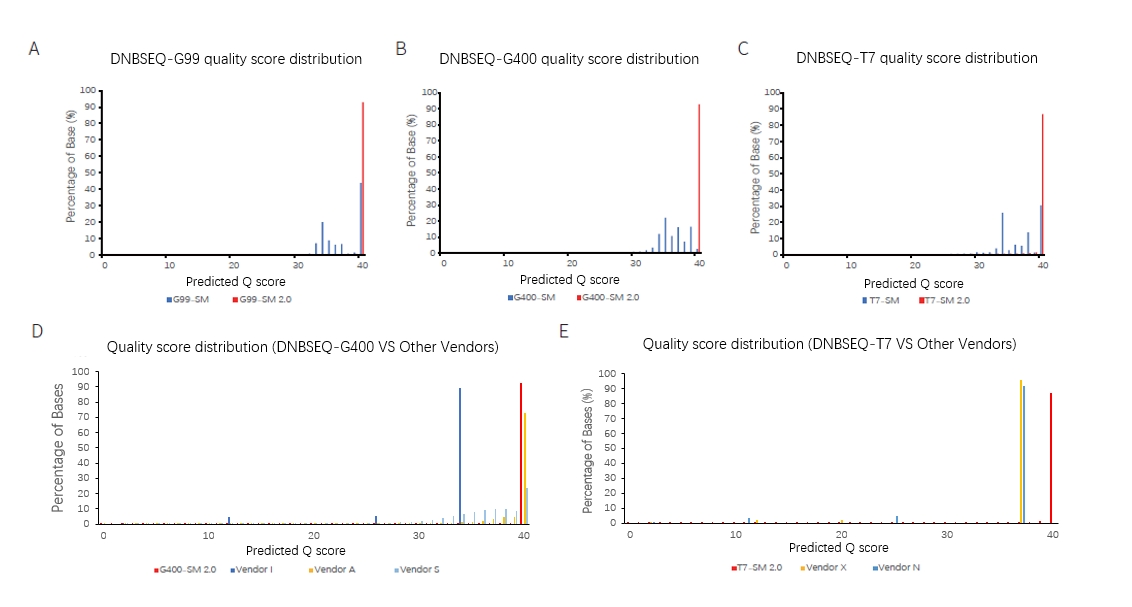
Figure 1. SM 2.0 could significantly improve on the sequencing quality of DNBSEQ platform. The distribution of bases at different Q scores obtained from DNBSEQ-G99(A), DNBSEQ-G400(B) and DNBSEQ-T7(C) combined with SM or SM 2.0. The comparison of data quality from DNBSEQ-G400(D) or DNBSEQ-T7(E) combined with SM 2.0 with other vendors. A is E. coli WGS library, and B~E are NA12878 WGS libraries.
Effectively improve the efficiency of variation detection in WGS research
The accuracy of SNP and InDel detection in DNBSEQ-G400 combined with SM 2.0 exceeded other vendors, reaching 99.8% and 99%, respectively (Figure 2A). Sequencing quality improvement obviously reduced data requirement for downstream applications. For example, the raw data required for reaching 30× average sequencing depth in WGS analysis is significantly lower than the industrial standard (120 Gb) or other commercial platforms (Figure 2B).
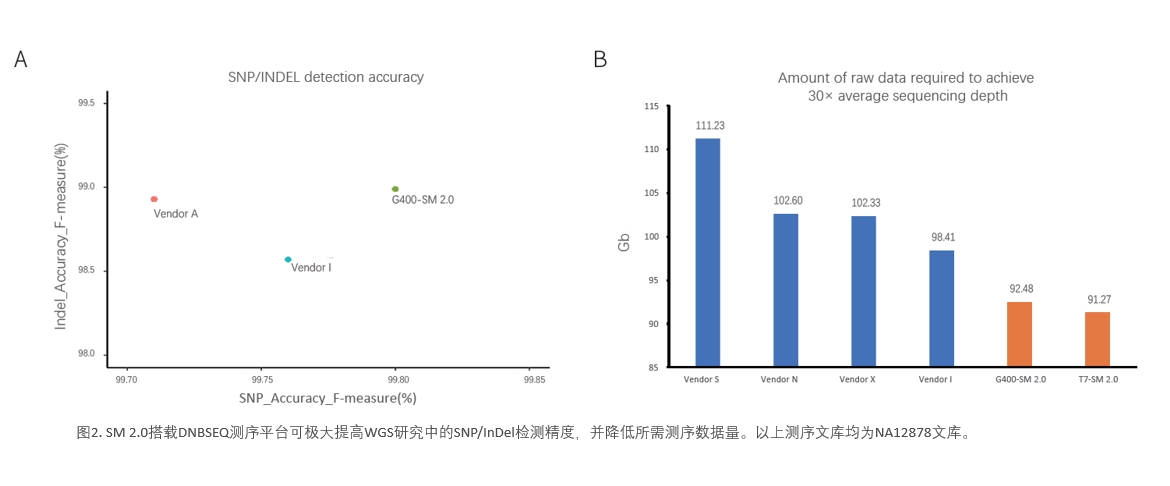
Figure 2. DNBSEQ platform combined with SM 2.0 can greatly improve the SNP/InDel detection accuracy and reduce data requirement in the WGS study. All the above libraries are NA12878 WGS
libraries.
Data source: Xpress genomics Sequencing
platform: DNBSEQ-G400
Application: Single-cell RNA sequencing (scRNA-seq)
Expanding the applications of single-cell sequencing
In the single-cell sequencing scenario, SM 2.0 combined with DNBSEQ-G400 have up to 90.88% bases’ sequencing quality reaching Q40, much better than SM. SM 2.0 could bring more total reads, higher mapping rate, lower mismatch rate, and more accuracy in detecting the expression level of specific allele.
Christoph Ziegenhai, CEO of Xpress genomics company, said: “We are excited to see the release of StandardMPS 2.0 sequencing reagent, which can improve the sequencing quality to Q40. Its early test data is very well and can help us to expand the applications of single-cell sequencing.”
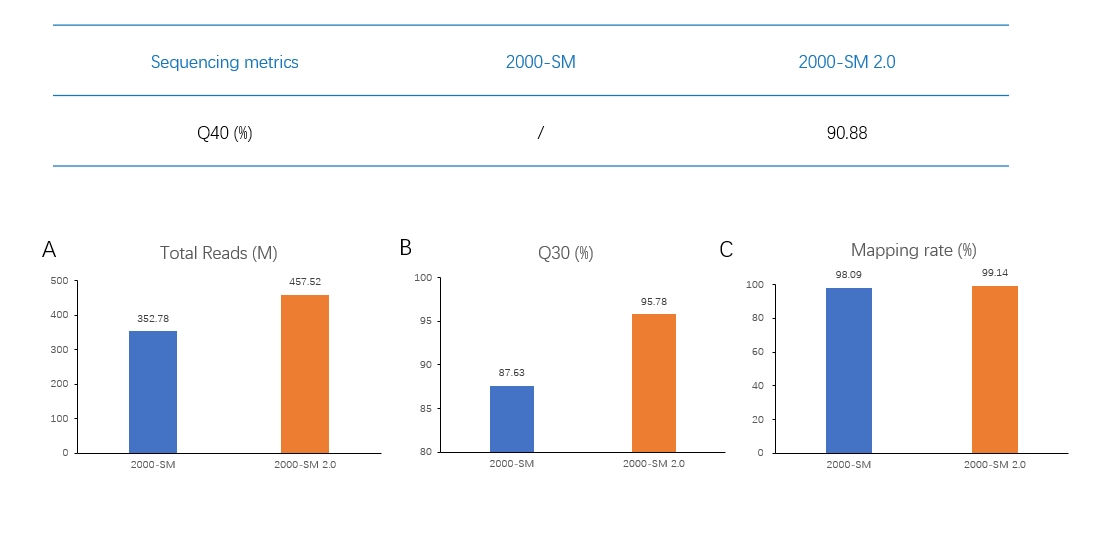
Data source: Ambiogen
Sequencing platform: DNBSEQ-T7
Applications: Microorganism research, metagenomic sequencing
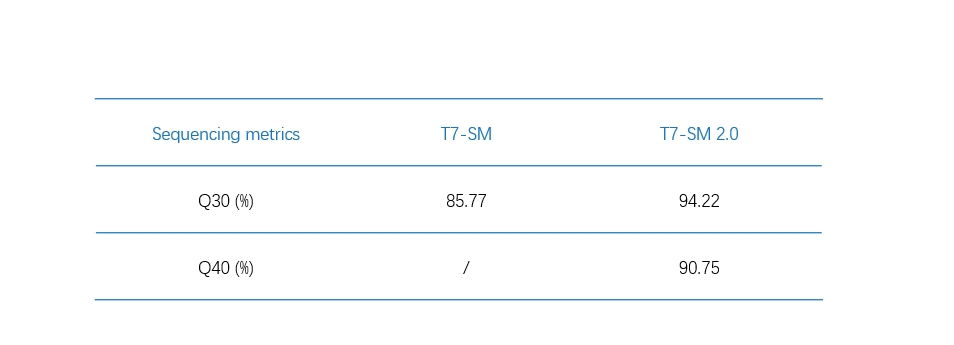
A powerful support for microorganism research
In the microorganism research, SM 2.0 combined with DNBSEQ-T7 can facilitate
Q40 to reach as high as 90.75% and Q30 up to 94.22%.
Maike Seifert, CEO of Ambiogen company highlights: “In the early test of metagenomic sequencing, StandardMPS 2.0 reagent showed an excellent Q40 sequencing quality. It makes sequencing data highly accurate and reliable, and can help us to analyze and study the complicated microbial communities more accurately.”
Conclusion
StandardMPS 2.0 sequencing reagent developed by MGI has been proven to assist DNBSEQ
sequencing quality reaching Q40 (>85%). Various applications, such as WGS, single-cell and microorganism studies, can benefit from the improvement of sequencing data quality. StandardMPS 2.0 combined with DNBSEQ platform can further accurately identify the low-frequency mutations and reduce the sequencing cost. These remarkable features facilitate in-depth analysis of complex biological processes within organisms, thereby significantly advancing the widespread adoption of high-throughput sequencing technology across various fields including oncology, infectious diseases, genetic disease diagnosis, and beyond.
StandardMPS 2.0 sequencing reagent has now been released globally and utilized in many sequencing platforms such as DNBSEQ-T7, DNBSEQ-G99, and DNBSEQ-G400 (this product is sold in China under the name of MGISEQ-2000). Should you have any questions, please reach out to your local sales staff.
References:
1. Secomandi S, Gallo G R, Sozzoni M, et al. A chromosome-level reference genome and pangenome for barn swallow population genomics[J]. Cell reports, 2023, 42(1).
2. Singh R R. Next-generation sequencing in high-sensitive detection of mutations in tumors: challenges, advances, and applications[J]. The Journal of Molecular Diagnostics, 2020, 22(8): 994-1007.
3. Beg A Z, Khan A U. Exploring bacterial resistome and resistance dessemination: an approach of whole genome sequencing[J]. Future Medicinal Chemistry, 2019, 11(3): 247-260.
4. Huang H, Kai Z, Wang Y, et al. Evaluating personalized circulating tumor DNA detection for early‐stage lung cancer[J]. Cancer Medicine, 2023.
5. Elazezy M, Joosse S A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management[J]. Computational and structural biotechnology journal, 2018, 16: 370-378.
6. Kaewprasert O, Tongsima S, Ong R T H, et al. Optimized analysis parameters of variant calling for whole genome-based phylogeny of Mycobacteroides abscessus[J]. Archives of Microbiology, 2022, 204(3): 190.
7. Pontes A, Hutzler M, Brito P H, et al. Revisiting the taxonomic synonyms and populations of Saccharomyces cerevisiae—phylogeny, phenotypes, ecology and domestication[J]. Microorganisms, 2020, 8(6): 903.



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













