The genotype imputation is a basic approach to impute the unobserved genotypes in the data from the genotyping arrays or genome sequencing, which can critically improve the fine mapping of variants in genome-wide association studies.
Based on large-scale and specific populations, the haplotype reference panel constructed by whole-genome sequencing data is a basic tool for genotype imputation. However, the main reference panels include databases such as 1000 Genomes Project Phase 3 (1KGP3), Haplotype Reference Consortium (HRC), and Trans-Omics for Precision Medicine (TOPMed) programs exhibited poor applicability and performance in the genotype imputation for East Asian populations. Currently, the multi-ethnic 1KGP3 reference panel is most commonly used for the genetic cohort studies of Chinese populations.
Therefore, the haplotype reference panel for Chinese and other East Asian populations constructed by the in-depth whole-genome sequencing data based on large-scale and representative populations achieves high-precision genotype imputation, which is of great value to the genomics studies in Chinese populations and disease cohorts.
High-throughput sequencing platform: advancing genotype imputation studies for populations
The Weiqing wang/Yanan Cao team from Ruijin Hospital (affiliated with the School of Medicine, Shanghai Jiao Tong University) published a paper titled The ChinaMAP reference panel for the accurate genotype imputation in Chinese populations in Cell Research on September 6, 2021. This study conducted a meta-analysis of the in-depth whole-genome sequencing data of 10,155 individuals from different regions and ethnic groups in China in the China Metabolic Analytics Project (ChinaMAP) to construct the largest ChinaMAP reference panel so far and build the online genotype imputation analysis tool platform “ChinaMAP Imputation Server” (already launched at http://www.mbiobank.com). This study can provide accurate and powerful reference data and reference panels for genotype imputation studies in Chinese and other East Asian populations.

It is noted that this study obtained basic data from MGI’s DNBSEQ sequencing platform and constructed a high-quality Chinese population database based on study cohorts covering various regions in China and conducted in-depth whole-genome data and phenotype analysis through the Chinese’s own instruments, platforms, and analytical methods, providing the powerful basis for study and prevention of disease mechanism, genetic counseling, and public health management.
ChinaMAP Imputation Server
Providing accurate reference data for Chinese genotype imputation studies and population-based disease studies
For a long time, many studies on genetic diseases in China have directly cited the data and conclusions from foreign countries. However, due to the huge differences in historical origins and genetic backgrounds between different populations and races, it is imperfect and unreliable to directly use biased knowledge and conclusions from other populations as the basis for disease risk assessment, genetic counseling, or diagnosis and treatment of Chinese populations.
Genomics and multi-omics big data from large-scale population cohorts are also playing a leading role in the prevention, diagnosis, and new drug development for major chronic diseases, tumors, and genetic diseases, and driving the reform in precision medicine for personalized health management as well as disease diagnosis and treatment.
This study was conducted by team members from Ruijin Hospital (affiliated with the School of Medicine, Shanghai Jiao Tong University), National Key Scientific Infrastructure for Translational Medicine (Shanghai), National Research Center for Translational Medicine of Shanghai Jiao Tong University, and SJTU Innovation Research Center, and will provide accurate reference data for Chinese genotype imputation studies and population-based disease studies.
In this study, the ChinaMAP reference panel contained 59.01 million genetic polymorphisms, and compared with the major reference panels of TOPMed, gnomAD, dbSNP, and 1KGP3, the ChinaMAP reference panel contained 30.24 million specific single nucleotide polymorphisms (SNPs), providing an important basis for the findings of novel variants in genomic studies of Chinese populations. In the imputation analysis of mimic and actual genotyping datasets for the Chinese populations, the ChinaMAP reference panel exhibited optimal imputation accuracy, precision, and sensitivity compared with 1KGP3, HRC, and TOPMed, significantly increasing the number of low-frequency loci such as loss-of-function loci and the coverage of common variants (mutation frequency >5%). Using the ChinaMAP Reference Panel for imputation of MAPCGA genotyping array data specific to Chinese populations, the coverage of loci with frequencies above 0.5% in the ChinaMAP database can reach more than 83%, showing the superiorities in Chinese genotype imputation studies of the reference panel constructed based on the representative ChinaMAP in-depth whole-genome sequencing database of Chinese populations.
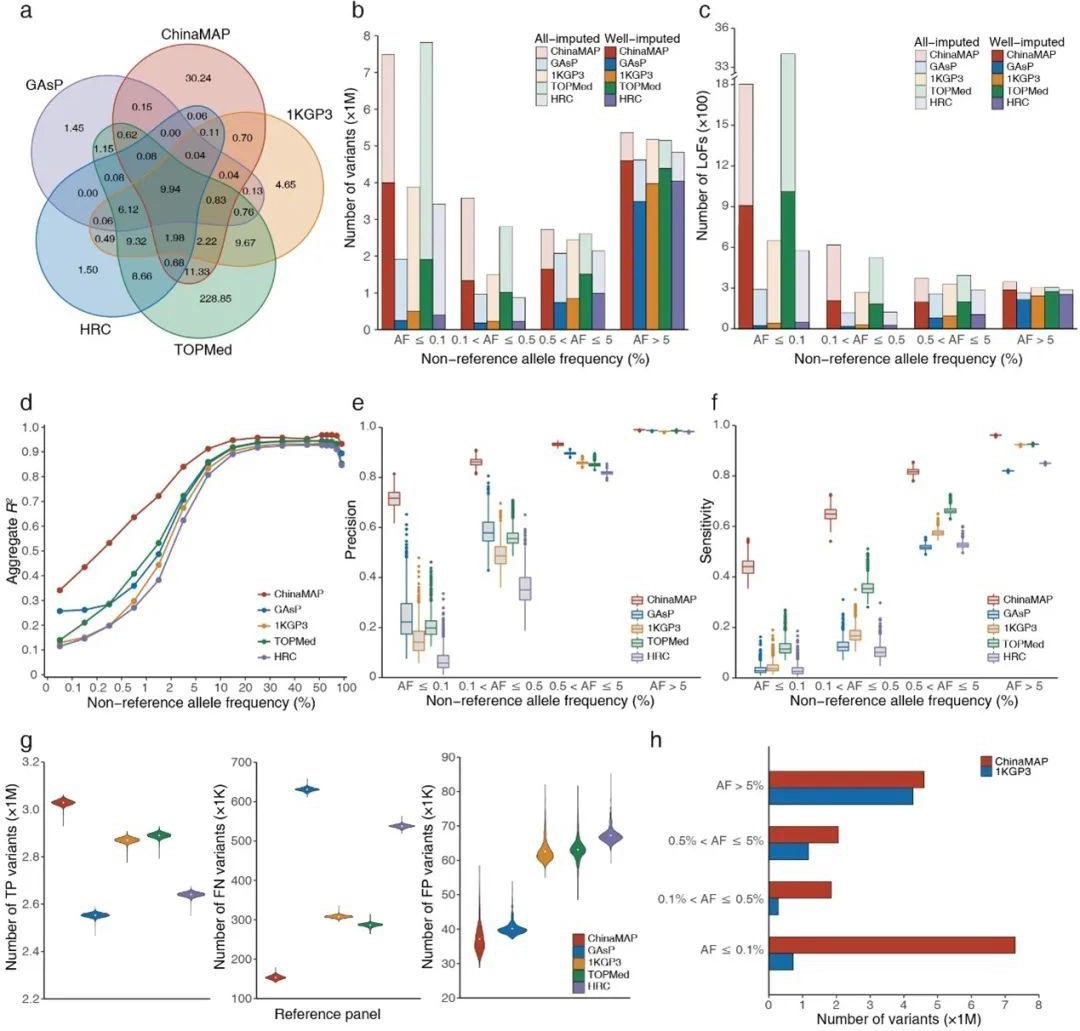
ChinaMAP reference panel genotype imputation performance
In addition to the data based on the domestic sequencing platform of MGI, this study also completed the construction and analysis of the ChinaMAP reference panel by means of a π 2.0 cluster supported by the Center for High-Performance Computing at Shanghai Jiao Tong University. ChinaMAP’s largest Chinese population reference panel and online imputation analysis tools have shown improved performance and contributed to population cohort genomics studies and precision medicine studies in China.
*In this study, Drs. Lin Li, Xiaohui Sun, Siyu Wang, and Peide Huang were the co-first authors.



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













