Today marks the International Day for Biological Diversity, a crucial reminder of the importance of biodiversity for the health of our planet and the well-being of all living beings. Biodiversity plays a vital role in maintaining ecological balance and supports the services that we rely on—clean air, water, food, and so much more. Research on environment and biodiversity explores the interrelationship between environmental composition, function, and biodiversity, as well as the impact of human activities on natural systems.
Interestingly, even the least human-reached regions of our planet - such as the Abyssal zones of the ocean and the icy expanse of Antarctica, are filled with secrets waiting to be uncovered and connected with human beings. These areas, often overlooked, hold clues to the history of life on Earth and help us understand the evolution of species over thousands of years.
Thanks to the advancements in environmental DNA (eDNA) - a useful tool to decrypt biodiversity levels in the environment, today we can easily identify the species that once inhabited these areas simply by collecting a cup of seawater, a handful of soil, or a tube of sediment.
In this article, we share 5 eDNA studies published in top journals that mainly explore the biodiversity in extreme areas including Abyssal zones and Antarctica. Empowered by MGI’s DNBSEQ™ technology, these studies reveal the adaptation mechanism and evolution history of species to extreme environments, providing invaluable scientific insights into sustainable development of nature resources and biodiversity conservation.
Study 1: Microbial ecosystems and ecological driving forces in the deepest ocean sediments

Journal: Cell
Publication Date: 2025
Method:16S rRNA gene amplicon sequencing and metagenomic sequencing
Platform: DNBSEQ-T series platform
Summary: The research team explored the abyssal environments of the Mariana Trench, Yap Trench, and Philippine Sea Basin at water depths of 6,000-11,000 meters, where the pressure reaches 1,100 atmospheres. They collected over a thousand samples, including water, sediment, macroorganisms, and rocks. Metagenomic sequencing of the sediment samples identified 7,564 prokaryotic microbial species, 89.4% of which were previously unreported. The study found that environmental selection pressures, such as high hydrostatic pressure, low temperature, and nutrient limitation, play a key role in shaping deep-sea microbial communities. It also revealed various mechanisms of deep-sea microbial adaptation to extreme environments, such as enhanced antioxidant mechanisms and the accumulation of intracellular compatible solutes, which are ubiquitous across different biological domains. The database generated by this research provides valuable resources for future studies on deep-sea microbial adaptation and has potential applications in biotechnology and environmental science.
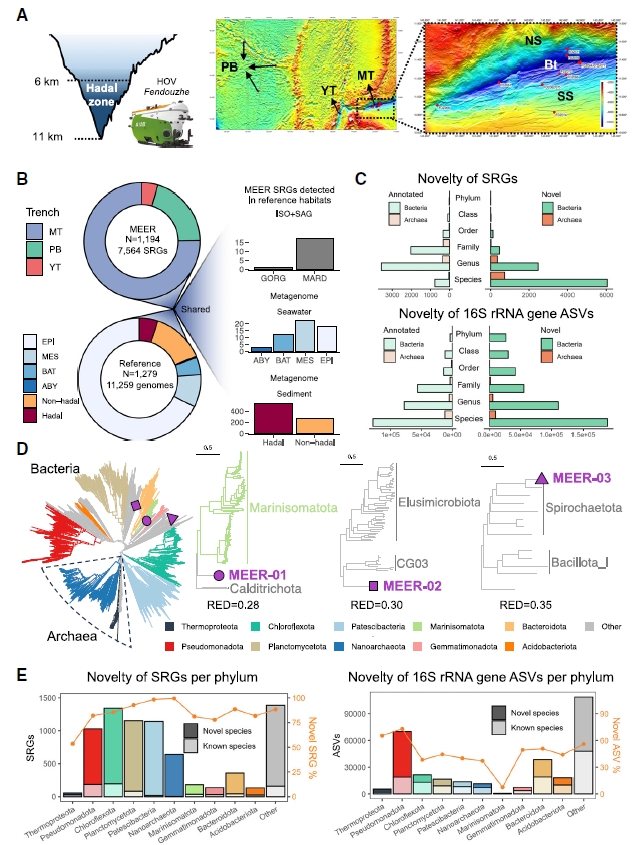
Study 2: The amphipod genome reveals population dynamics and adaptations to hadal environment

Journal: Cell
Publication Date: 2025
Method: Whole-genome sequencing
Platform: DNBSEQ-G400, DNBSEQ-T10
Summary: The research team collected amphipod samples from the Mariana Trench, Yap Trench, and Philippine Sea Basin and performed whole-genome sequencing, successfully assembling a high-quality chromosome-level genome of the amphipod (13.92 Gb). Comparative genomic analysis revealed no genetic differentiation among amphipod populations from different depths (approximately 7,000-11,000 meters) in the Mariana Trench, indicating a fully mixed population capable of adapting to varying hydrostatic pressures. However, significant genetic differentiation was observed between the amphipod populations in the West Philippine Basin and those in the Mariana and Yap Trenches, likely due to geographical isolation. The study also found that the synergistic relationship between amphipods and their symbiont Psychromonas may be key to adapting to abyssal environments. This research challenges traditional pressure resistance theories and highlights the central role of symbiotic systems in extreme survival. Its findings not only expand the frontiers of life sciences but also provide a scientific basis for deep-sea ecological protection and resource sustainable utilization.

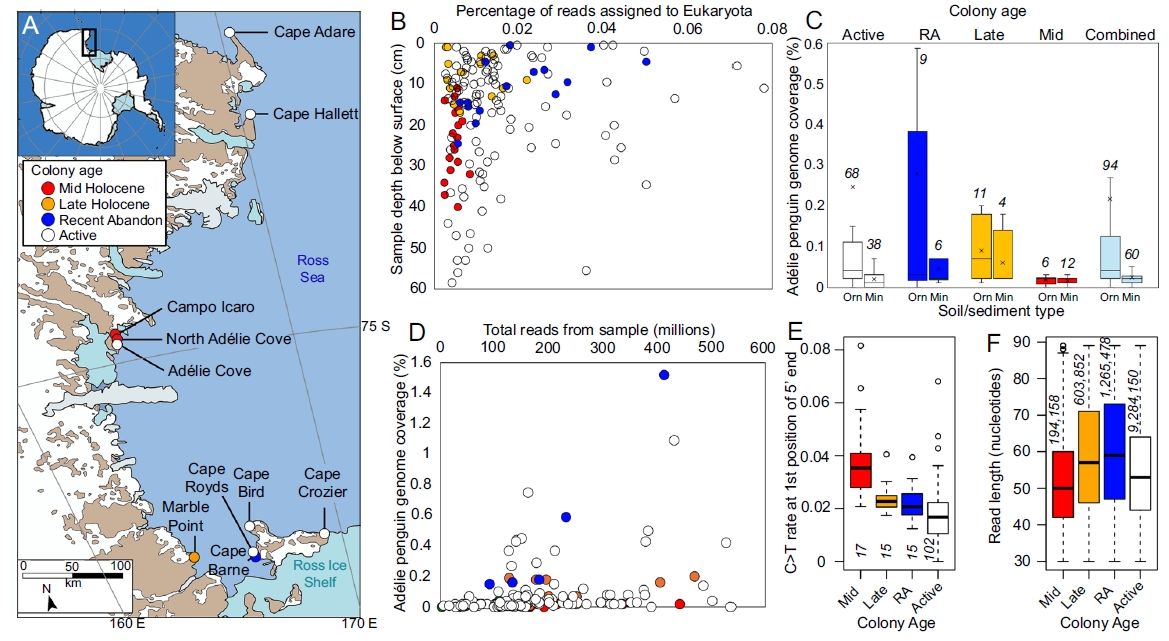
Study 3: Sedimentary DNA insights into Holocene Adélie penguin (Pygoscelis adeliae) populations and ecology in the Ross Sea, Antarctica

Journal: Nature Communications
Publication Date: 2025
Method: Metagenomic sequencing
Platform: DNBSEQ-T series platform
Summary: The study analyzed 156 sediment samples from six active and four abandoned Adélie penguin colonies from Ross Island and the East Victoria Land coastline, the samples spanned 6,000 years. Metagenomic sequencing of these samples, combined with the lowest common ancestor (LCA) analysis method, identified DNA sequences of animals, plants, fungi, and microbes. The research revealed population changes in Adélie penguins, including their distribution and numbers across different historical periods. It also detected the presence of other local species, such as various birds, seals, and invertebrates. For instance, the DNA of southern elephant seals was detected at Cape Hallett, indicating it was once a breeding ground for them. This study opens a window to discover the evolutionary history of Antarctic ecosystems over millennia, deepening the understanding of Adélie penguins' adaptive strategies and highlighting the power of eDNA technology in unraveling the complex relationships between "climate, ecology, and evolution." The findings provide key scientific evidence for Antarctic biodiversity conservation and global change biology.
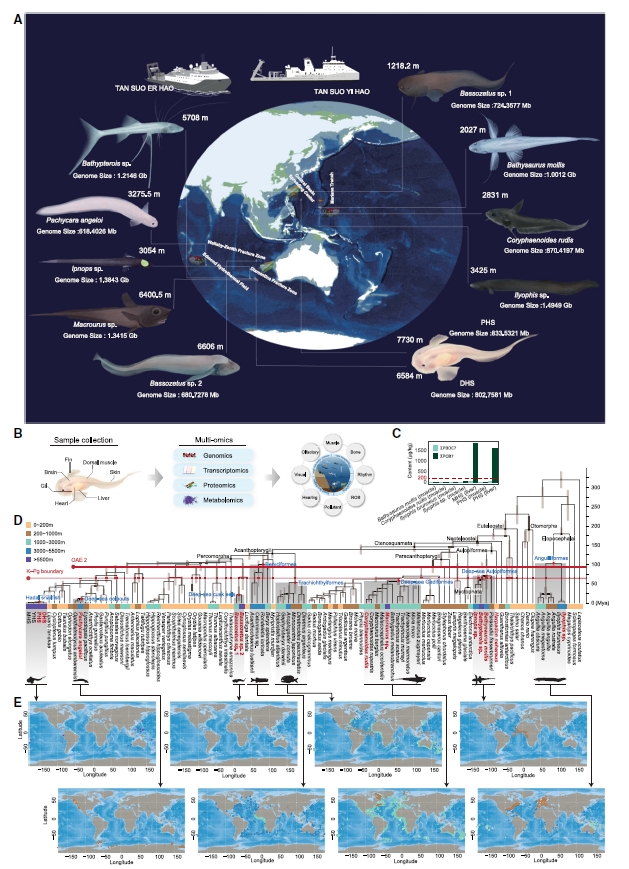
Study 4: Evolution and genetic adaptation of fishes to the deep sea

Journal: Cell
Publication Date: 2025
Method: DNA barcode sequencing, transcriptome sequencing
Platform: DNBSEQ-G400
Summary: The research team conducted extensive sampling across the western Pacific to the central Indian Ocean, generating genome for 12 species, including 11 deep-sea fish species. Comparative genomic analysis of the fish hosts and their phototrophic symbionts across different environmental, revealed various mechanisms of deep-sea fish adaptation to extreme environments. For example, a highly conserved convergent mutation in the rtf1 gene of most deep-sea fish living below 3,000 meters was found to enhance transcription efficiency, likely a favorable adaptation to deep-sea environments. The study also challenged the previous assumption of a linear relationship between TMAO content and depth. While TMAO levels in fish do increase with depth from 0 to 6,000 meters, this trend does not continue beyond 6,000 meters. Additionally, the team detected high concentrations of persistent organic pollutants like polychlorinated biphenyls (PCBs) in fish from the Mariana Trench, indicating that these synthetic pollutants have penetrated the deepest parts of the ocean. This research reshapes our understanding of the limits of deep-sea life and sets new benchmarks for extreme environment biology through interdisciplinary technological integration. Its findings provide key scientific evidence for deep-sea ecological protection, biomedicine (such as the of application pressure-adaptive genes), and climate change research.
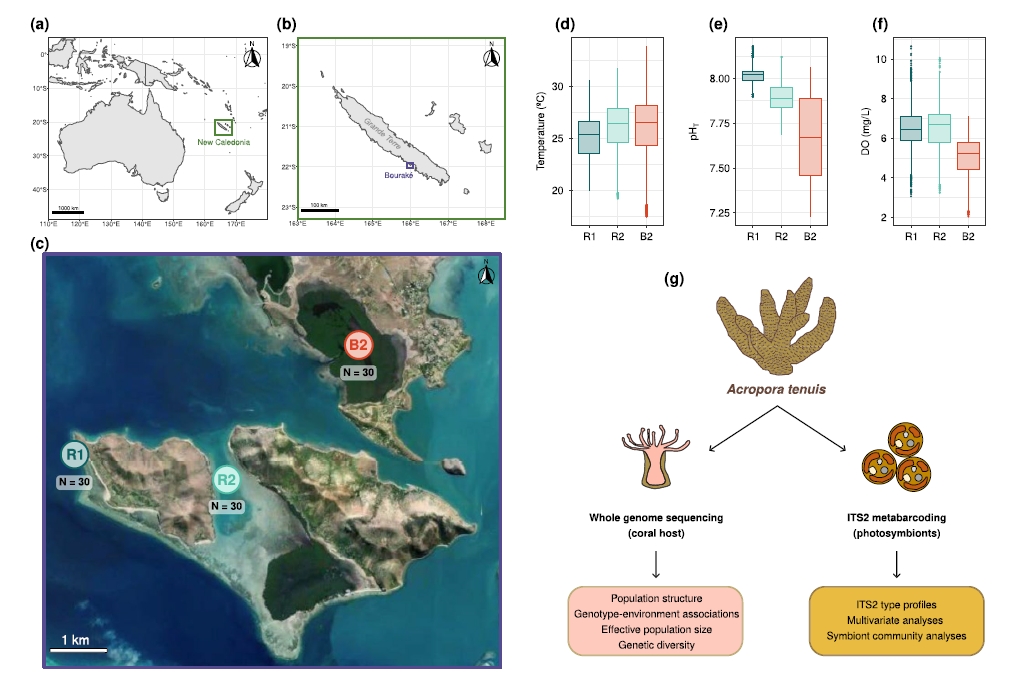
Study 5: Rapid Evolution in Action: Environmental Filtering supports Coral Adaptation to a Hot, Acidic, and Deoxygenated Extreme Habitat
Journal: Global Change Biology
Publication Date: 2025
Method: Whole-genome sequencing
Platform: DNBSEQ sequencing platform
Summary: The research team sequenced corals and their phototrophic symbionts from Bouraké Lagoon and two nearby control reefs. By analyzing gene variations and community structures of coral hosts and phototrophic symbionts across different environmental, to study explored coral adaptability. It found that corals' adaptability is reflected not only in their association with different phototrophic symbionts but also in specific genomic loci of the coral host. Corals can flexibly associate with various phototrophic symbionts to enhance their tolerance to extreme environments. This study first provided evidence of corals' rapid evolutionary capacity in response to multiple extreme environmental in natural ecosystems. It offers revolutionary insights into coral reef protection: filtered genotypes by environment, humans may actively intervene to accelerate corals' adaptation to climate change. This discovery not only brings hope for coral reef ecosystem restoration but also a classic case for understanding mechanisms of rapid biological evolution.
The DNBSEQ sequencing platform adopts unique DNBSEQ sequencing technology, featuring high accuracy, low repeat sequence error rates (<0.1%), and low cross-contamination risks. It is particularly suitable for detecting complex environmental samples such as soil and water eDNA, enabling precise analysis of genetic information from low-abundance species and supporting large-scale parallel sequencing to significantly reduce unit costs. Additionally, the DNBSEQ sequencing technology's high compatibility with low-quality/low-concentration DNA and its relatively long sequencing read length effectively support the analysis of complex microbial communities and the assembly of animal and plant genomes, becoming a core technological support for environmental and biodiversity research.

Recently, MGI eDNA metabarcoding sequencing package has been upgraded. It now includes a portable smart sampler to meet outdoor rapid sampling needs, offering a truly one-stop product solution from sampling to reporting. The new ATOPlex 16SV4 rDNA library prep kit has been added, combined with the DNBSEQ-E25 sequencer, can stably and quickly produce accurate data on board. The MetaSIS analysis software has also been upgraded to support customizable primer sequences and custom databases.
Adhering to the philosophy of "innovation-driven leadership in life sciences," MGI will continue to advance product iteration and updates, providing more efficient and cost-effective tools for biodiversity monitoring, endangered species monitoring, and invasive species early warning.



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













