The "Product FAQs" column collects products related questions which MGI customers inquiries frequently. And we organizes FAQs which will be released in the future to help you understand and use MGI products better.
Content:
Q1: What is the brand positioning of CycloneSEQ?
Q2: What is CycloneSEQ-WT02?
Q3: What is the sales mode of CycloneSEQ-WT02?
Q4: When and where does MGI launch the CycloneSEQ-WT02?
Q5: What components make up the CycloneSEQ-WT02 system?
Q6: What is the sequencing principle of CycloneSEQ-WT02?
Q7: What is the principle difference between CycloneSEQ and DNBSEQ?
Q8: CycloneSEQ-WT02 Performance Parameters
Q9: What is the technological strength of CycloneSEQ-WT02?
Q10: What are the applications of the combined CycloneSEQ and DNBSEQ sequencing?
Q11: Does CycloneSEQ have automated library preparation solutions?
Q12: What is CycloneSEQ-WY01?
Q13: What is the launch timeline for CycloneSEQ-WY01?
Q1: What is the brand positioning of CycloneSEQ?
CycloneSEQ is the nanopore sequencing technology platform, dedicated to developing a series of nanopore sequencers, flow cells, and reagent consumables. Based on the CycloneSEQ platform, we aim to provide customers an integrated "short and long read" sequencing solution. In addition to facilitating existing customers with the expansion of long-read sequencing capabilities, there is a stronger expectation to contribute to the further expansion of downstream sequencing applications. This sequencing platform has extensive application prospects in the field of life science. It can not only empower the cutting-edge scientific research but also significantly promote the development in various fields such as public health, environmental protection, precision medicine, etc.
Q2: What is CycloneSEQ-WT02?
CycloneSEQ-WT02, a nanopore gene sequencer launched in 2024, features a dual flow cell architecture that enables independent operation simultaneously. With flexible throughput options and exceptional capabilities such as rapid coverage, swift sequencing, and flexible testing. It effectively addresses the challenges of complex sequences, meeting the diverse sequencing needs across various applications, such as microbial genome research, microbial metagenomes, amplicon-based microbial detection, targeted disease testing, small whole-genome sequencing whole transcriptome study, etc.
Q3: What is the sales mode of CycloneSEQ-WT02?
MGI has acquired the global market distribution rights for nanopore sequencing equipment and related products such as reagents and consumables from Hangzhou CycloneSEQ Technology (with exclusive distribution rights in Mainland China, Hong Kong, Macau, and Taiwan, and non-exclusive rights in other countries and regions). The distribution period lasts until December 31, 2029, during which the CycloneSEQ-WT02 is sold on the MGI platform.
Q4: When and where does MGI launch the CycloneSEQ-WT02?
MGI serves as the distributor to sale the CycloneSEQ series of nanopore sequencing products according to the cooperation agreement with Hangzhou CycloneSEQ Technology on June 28, 2024. CycloneSEQ-WT02, as the first product in this series, was available for early trial customer delivery on June 28, 2024, and is scheduled for an official global launch on September 9 of the same year, with the China region being the first to market, followed by other overseas regions according to their local conditions.
Q5: What components make up the CycloneSEQ-WT02 system?
The CycloneSEQ-WT02 sequencer requires the CycloneSEQ WT sequencing flow cell, library preparation kits (such as the CycloneSEQ Universal Library Prep Set or CycloneSEQ 24 Barcode Library Prep Set), and the CycloneSEQ WT sequencing kit, etc. Additionally, a specialized high-performance computer is required. We recommend our CycloneSEQ workstation, which is already pre-installed with our CycloneMaster software that can perform all core sequencing functions from data acquisition and device control to real-time analysis and basecalling. Furthermore, the output data from CycloneSEQ-WT02 is compatible with mainstream third-party nanopore sequencing analysis software such as minimap2, clair3, flye, etc.
Q6: What is the sequencing principle of CycloneSEQ-WT02?
During the CycloneSEQ-WT02 sequencing process, the DNA library molecules linked to the motor protein are drawn to the vicinity of the nanopore protein embedded in the biomimetic membrane under the influence of electric field forces, where they are captured by the nanopore protein. Meanwhile, motor proteins situated near the nanopore proteins entrance, steadily and rapidly unwinding the DNA. This allows the DNA libraries to pass through the nanopore as a single strand. Different DNA bases and their arrangement impede the current to varying degrees, triggering the current fluctuations. The channel sensor captures these current fluctuation data and transmits them to the computer system, where basecalling algorithms parse the information to achieve real-time and accurate gene sequencing.
Q7: What is the principle difference between CycloneSEQ and DNBSEQ?
◾Signal recognition methods: The DNBSEQ platform relies on optical signal detection, either through excitation light mode (detecting fluorescence signals via an optical device) or self-luminous mode (detecting fluorescence signals through photoelectric conversion). In both cases, the detection is fundamentally fluorescence-based. While the CycloneSEQ platform bypasses the luminescence process entirely, generating an electrical signal directly from sequencing, which is then converted into a base sequence. This approach eliminates the need for large-scale photographic equipment.
◾Data Type: The data logic of the DNBSEQ platform is based on a single base per cycle as the data unit, meaning that all reads of uniform read length are generated at a fixed time. The sequencing read length is primarily related to the sequencing mode, such as SE100, PE150, etc. The CycloneSEQ platform uses individual reads as the data unit, and during sequencing, each sequencing channel performs real-time basecalling simultaneously as a read pass through the nanopore, obtaining the FASTQ information for each read. Therefore, the sequencing time also depends on the demand for the amount of sequencing data.
◾Read Length: The DNBSEQ platform ensures that read lengths are consistent across each cycle, while the read lengths within a CycloneSEQ platform run may vary. The read length is predominantly determined by the length of the initial sample library.
Q8: CycloneSEQ-WT02 Performance Parameters
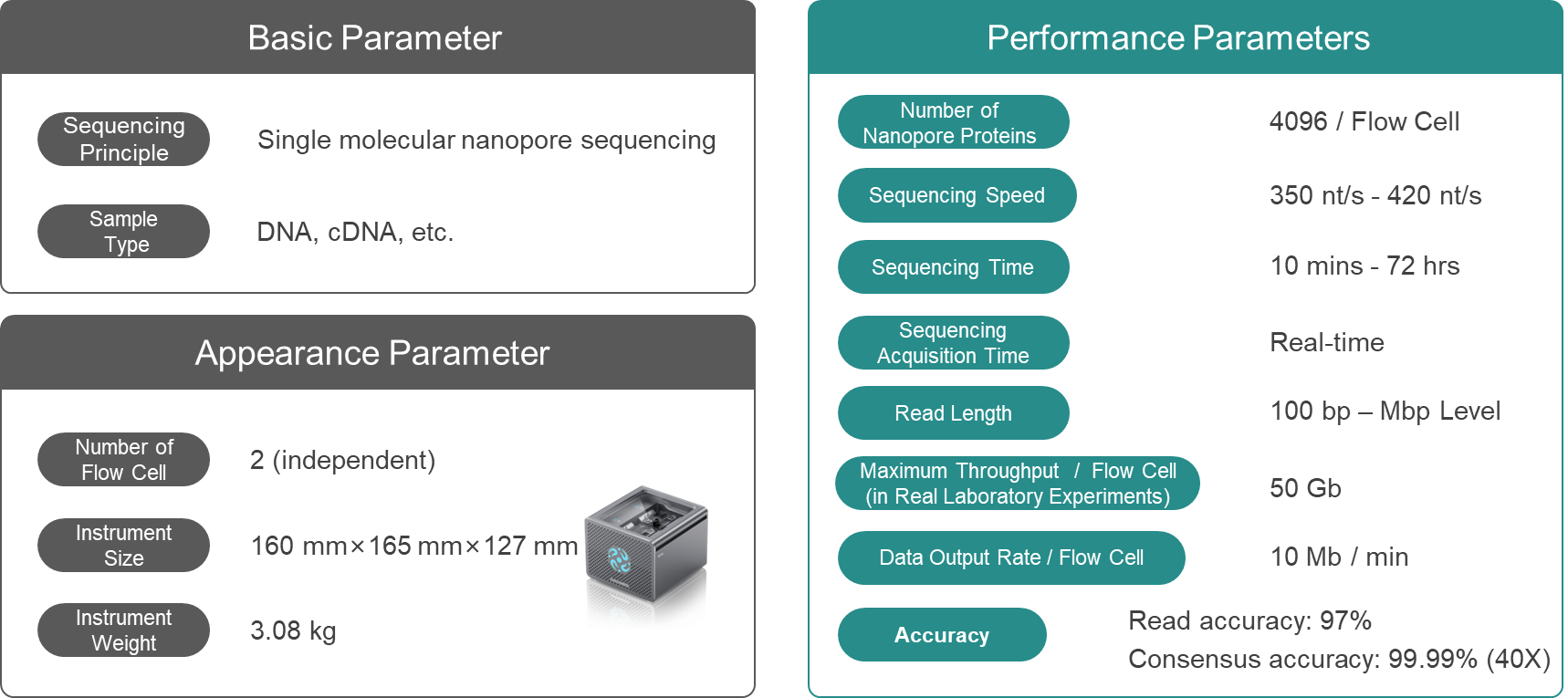
◾Sequencing Time: The CycloneSEQ-WT02 sequencing technology is a flexible start-and-stop mode, allowing run times to be set according to customer requirements, typically ranging from 10 minutes to 72 hours. (The longest sequencing run time in the laboratory is 107 hours.)
◾Sequencing Throughput: The sequencing throughput of CycloneSEQ-WT02 is flexible, with the actual output, per flow cell in the laboratory varying from 10 Mb to 50 Gb, determined jointly by the sequencing sample and the sequencing time. Currently, the highest actual data output per flow cell in the laboratory exceeds 50 Gb. In actual sequencing, the output may
vary with different sample types and applications.
◾Read Length: The read length is compatible with a range of 100 bp to Mbp. The read length is related to the sample conditions, N50 typically being 30~40 Kbp; some reads can reach the Mbp level. When using ultra-long sequencing operation methods, the read length can be increased to hundreds of Kbp or more. (The longest read length sequenced in the laboratory is 1.1 Mbp)
◾Sequencing Accuracy: The mode accuracy of raw sequencing is Q15 (97%). There are slight differences in quality values under different sample and experimental conditions, with the accuracy rate of a single sequencing run ranging from 94% to 97%. When the sequencing depth reaches 40X, the consensus accuracy is Q40 (99.99%).
Q9: What is the technological strength of CycloneSEQ-WT02?
Protein engineering, flow cell design, chemical reactions, and basecalling algorithms are the main components of CycloneSEQ nanopore sequencing technology. The technological competition in nanopore sequencing is currently focused on these four components:
◾Proteins: In the analysis of over 500 million deep-sea metagenomic databases collected from extreme environments such as the Mariana Trench, it was discovered that the deep sea has a high abundance of proteins related to genetic manipulation, with greater structural diversity. Among them, there is a highly robust nucleic acid manipulation protein, whose core
functional region structure is significantly different from most known motor proteins, with stronger unspinning ability, and it is a source of hydrothermal region and has high thermal stability. Another is a densely structured pore protein formed by the self-assembly of homopolymers, characterized by strong structural rigidity, low sequencing noise, and high quality, with a small pore diameter. Through protein engineering, the motor protein has been designed to have fast sequencing speeds, strong stability, and the ability to sustain sequencing over long periods, while the pore protein features strong capture, high stability, sustained sequencing, and high signal-to-noise ratios, forming the core protein foundation of CycloneSEQ.
◾Flow cell: A single flow cell can have up to 4096 nanopore protein sensors, arranged in a high-density array. The sensor microcavity diameter and spacing have been finely optimized for optimal membrane pore fitting, achieving ultra-low sequencing noise. The expanded microcavity volume allows for ultra-long sequencing durations. The integration of Bio Micro
Electro Mechanical System (BioMEMS) with Application-Specific Integrated Circuits (ASIC) enables the precise detection of picoampere-level currents, providing real-time and stable reading of nucleic acid sequence information.
◾Chemical Reaction Systems: The chemical environment near the nanopore is crucial for obtaining high-quality sequencing data. Unlike traditional large-scale chemical reactions, CycloneSEQ has developed a new type of chemical reaction, using the electronic flow cell of each pore to control ion concentration. By shielding all magnesium ions in the buffer and only
releasing them in a very small area near the pore using voltage, ATP-Mg is activated, thereby controlling the activity of the motor protein.
◾Algorithms: Incorporating state-of-the-art (SOTA) solutions from the field of Automatic Speech Recognition (ASR), the sequencing decoding algorithm utilizes a deep neural network trained on vast amounts of data to identify the optimal sequence from all possible combinations and decodes it with precision. The CycloneSEQ has independently developed a basecalling
algorithm model. This algorithm model, based on extensive data and large-scale distributed training, achieves a high accuracy rate of 97% and is capable of real-time sequencing decoding for multiple flow cells, demonstrating robust performance.
Q10: What are the applications of the combined CycloneSEQ and DNBSEQ sequencing?
These two technologies can perfectly complement each other in terms of sequencing length and accuracy, enabling more comprehensive and precise analyses in various applications, such as:
◾ Genome Assembly: Short-read sequencing technology provides high coverage and accuracy, while long-read sequencing offers continuous long sequence segments. The combination of these two can lead to the assembly of high-quality genomic sequences.
◾Detection of Genomic Structural Variations: Short-read sequencing offers accurate detection of SNPs and INDELs, while long-read sequencing directly detects complex structural variations, providing more precise genomic information.
◾Transcriptome Research: Short-read sequencing is used for quantitative expression analysis, and long-read sequencing retrieves full-length transcript sequences, providing a more detailed view of the transcriptome.
Q11: Does CycloneSEQ have automated library preparation solutions?
An automated library preparation instrument adapted to the CycloneSEQ platform has been developed and it is scheduled for an official launch in the first quarter of 2025. Meanwhile, we are also developing adaptations to other MGI's existing automated library preparation instruments.
Q12: What is CycloneSEQ-WY01?
CycloneSEQ-WY01 nanopore gene sequencer is scheduled for an official launch in 2025. Positioned as a high-throughput nanopore gene sequencer in the CycloneSEQ platform, it is equipped with a high-density flow cell and ultra-high throughput capabilities, allowing for ultra-long read lengths, real-time sequencing, and continuous testing. Its outstanding performance indicates extensive applicability in diverse applications, including large genome sequencing, human genome resequencing, genome assembly, full-length transcriptome sequencing, and epigenome sequencing, etc.
Q13: What is the launch timeline for CycloneSEQ-WY01?
The official launch of CycloneSEQ-WY01 is scheduled for the first half of 2025.




 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













