Tuberculosis, caused by Mycobacterium tuberculosis (M. tb), and AIDS caused by the human immunodeficiency virus (HIV), are two formidable infectious diseases. While one is bacterial and the other viral, they can synergize to inflict a devastating impact on patients.According to the latest data from the World Health Organization (WHO),tuberculosis alone causes 1.3 million deaths in 2022,with 167,000 of these fatalities occurring among HIV-positive individuals. The co-infection significantly elevates the risk of developing tuberculosis, with HIV-positive individuals facing a 20-fold increased susceptibility. In the same way, Mycobacterium tuberculosis infection exacerbates the symptoms in people with AIDS1.
Over time, patients with tuberculosis (TB) and HIV may be at risk of increased pathogen resistance. Considering that, World Health Organization (WHO) has identified antimicrobial resistance as one of the ten most significant global public health challenges. To tackle this challenge, targeted sequencing technology based on massively parallel sequencing (MPS) has demonstrated outstanding advantages in microbial traceability and the discovery of drug resistance genes. Itprovides more comprehensive variant information while balancing detection specificity and sensitivity compared to traditional molecular testing methods.
MGI ATOPlex multiplex PCR library preparation platform is based on ultra-high-plex PCR technology, capable of target amplification of multiple target regions at the same time, with a customizable panel, low requirements for input DNA integrity, amplification in one tube and anti-pollution system. This platform enables high-quality target sequencing when paired with MGI sequencers and automation platforms.
In the field of public health, MGI has launched ready-to-use full-flow sequencing combination products based on the ATOPlex multiplex PCR library preparation platform for a variety of infectious diseases such as COVID-19, influenza A and B, monkeypox anddengue fever. Recently, MGI has launched two new targeted amplification sequencing combination products for HIV and Mycobacterium tuberculosis, comprehensively covering the whole process of library preparation to sequencing and analysis for HIV andM. tb,and their drug resistance genes.

ATOPlex Technical Characteristics
HIV-1 Drug Resistance Sequencing Products Package
Product introduction
Based on DNBSEQ-G99, DNBSEQ-E25 and DNBSEQ-G50 sequencing platforms,MGIrecently officially launched the HIV-1 drug resistance sequencing product package, which empowers the prevention and control of AIDS with gene technology. Based on ATOPlex library preparation reagents and the self-developed automated sample preparation system and analysis software, the product package covers the entire process of library preparation, sequencing and report output, and is capable of rapid and accurate drug resistance detection and typing of HIV-1 virus in samples.
Product Highlights
Low-Frequency Resistance Detection:Utilizes deep sequencing to detect resistance with a mutation frequency of less than 20%, a level undetectable by Sanger sequencing.
Automation Friendly:Compatible with the MGISP-100/MGISP-960 Automated Sample Preparation System, and HIV-GenomePro platform, enabling automated library preparation and data analysis, thus saving labor.
Various Analysis Functions:The HIV-GenomePro software offers virus typing, gene assembly, mutation detection, and evolutionary traceability analysis. It accurately predicts and annotates drug resistance and can merge multi-sample sequences for HIV-trace analysis to construct the molecular network.

Whole workflow ofHIV-1 Drug Resistance Sequencing Product Package
Testing data
RNA extracted from plasma and cultured virulent strains was used as samples for library preparation. These samples were sequenced and analyzed using the DNBSEQ-G99ARS platform and the results were compared with those obtained from Sanger sequencing.
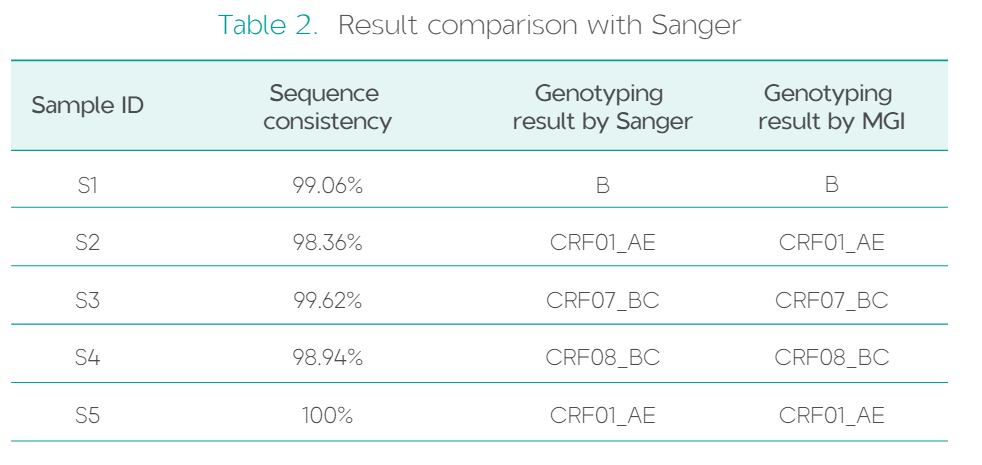
High consistancy with Sanger sequencing results
Sequencing results obtained using the HIV-1 Drug Resistance Sequencing Products Package showed>98% sequence identitywith those obtained from the Sanger sequencing protocol.Additionally, the typing results were generally consistent between the two methods.
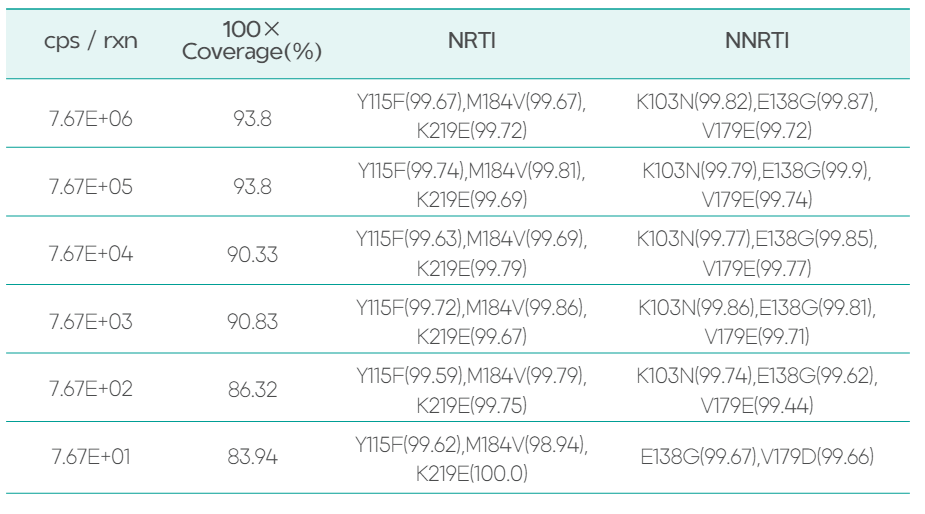
Work for detecting low-frequency mutations
The HIV-1 Drug Resistance Sequencing Products Package detectslow-frequency mutations (red-marked resistance sites) that are not detected by the Sanger sequencing protocol, with mutation frequencies in parentheses.

The RNA extracted from cultured strains was used as samples (reference resistance sites: Y115F, M184V, K219E, K103N, E138G, V179E), which were subjected to concentration-gradient dilution. Subsequently, library preparation, sequencing, and analysis were performed. All reference resistance sites were accurately detected when the viral copy number in the reaction was 7.67E+02. However, the K103N resistance site was failed to be detected when copy number was as low as 7.67E+01. Therefore, the lowest detection limit was as low as 102cps/rxn. The mutation frequencies of each mutation site were consistent across different concentration gradients, indicating excellent reproducibility.
Comprehensive Analysis Capabilities
The HIV-GenomePro analysis software provides comprehensive functionalities, including gene assembly, precise drug resistance locus annotation, and detailed phylogenetic analysis.
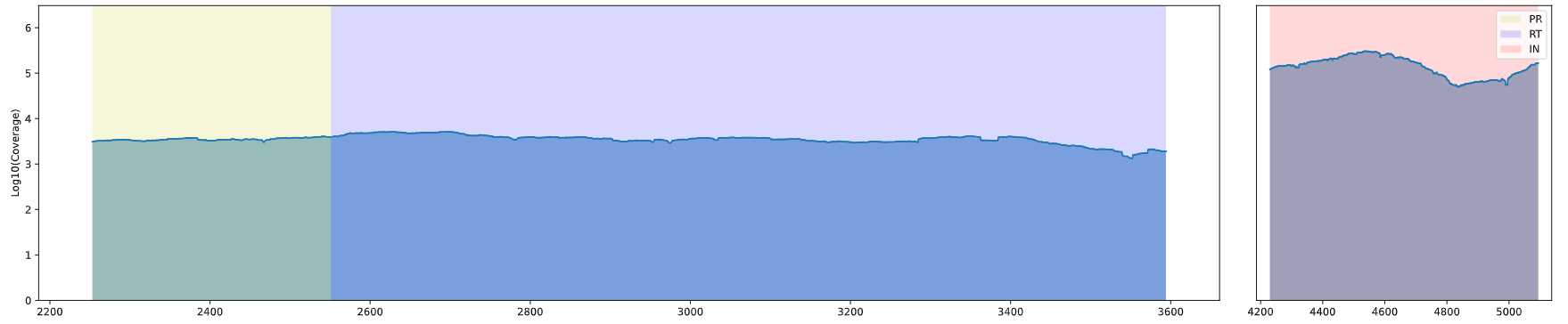
Gene Depth Coverage Map, "Depth Coverage Map" shows the depth coverage of reads for each chromosome/genome segment, with the X-axis being the base coordinates of the chromosome/genome segment, and the Y-axis being the lg-transformed value of the depth of the reads.
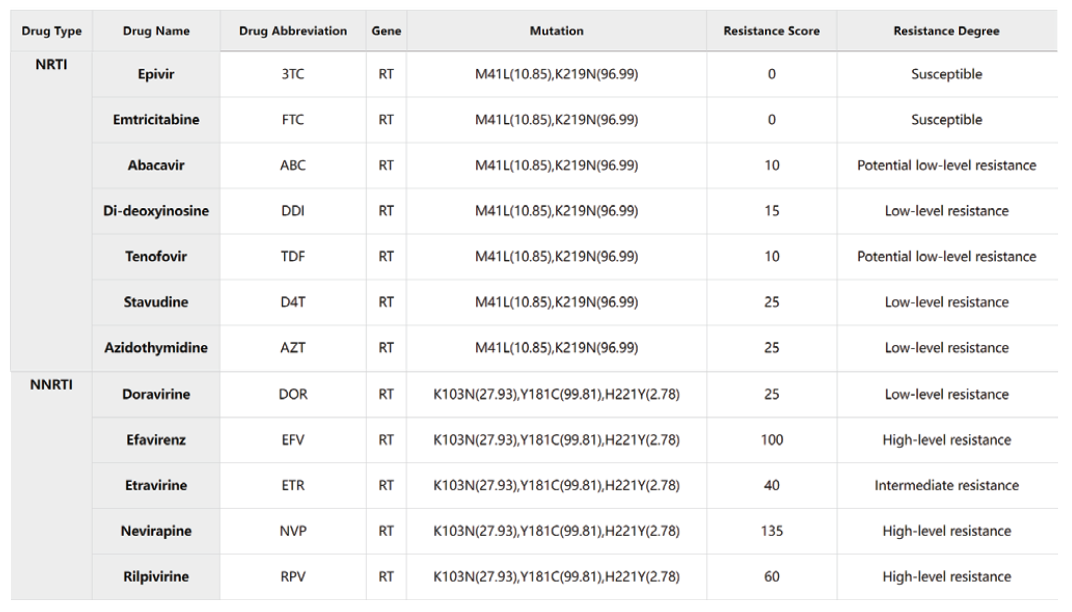
Results of drug resistance analysis
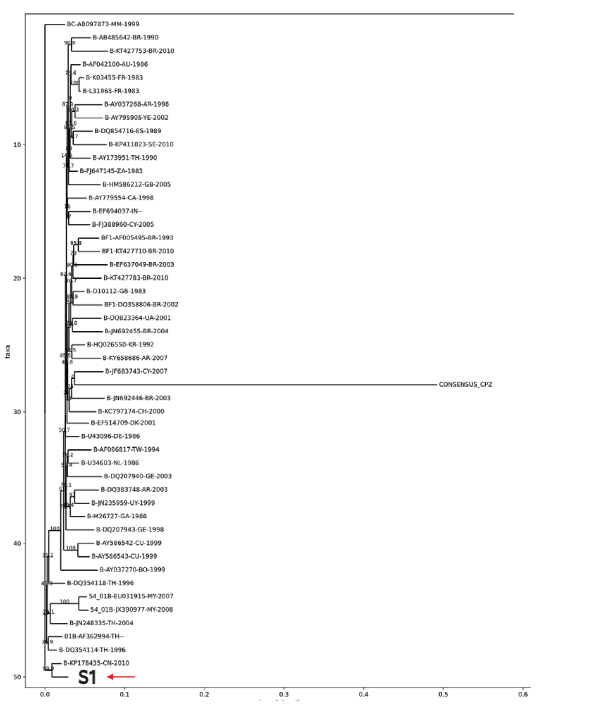
Evolutionary cladogram
ATOPlex Mycobacterium Tuberculosis Targeted Sequencing Package
Product introduction
Leveraging ATOPlex multiplex PCR technology, MGI has launched ATOPlex MTB library preparation kit. Combined with DNBSEQ sequencing platform, the kit can perform rapid and accurate targeted sequencing of drug resistance and identification regions of Mycobacterium tuberculosis. Addionally, MTB-Explorer software automatically analyzes Mycobacterium tuberculosis complex (MTBC) and mycobacteria for typing, mutation detection, drug resistance prediction, phylogenetic analysis, and generating comprehensive reports. The ATOPlex Mycobacterium Tuberculosis Targeted Sequencing Package encompsses the entire process from library preparation to sequencing and report output, providing robust support for public health prevention and control.
Product Highlights
Suitable for culture-free sample: This combination products supports mixed sample input such as sputum and lavage without the need for microbial culture.
Easy operation for library preparation: Utilizing the ATOPlex multiplex PCR library preparation platform, target area amplification and library preparation are achieved with a simple 2-step amplification process in a single tube.
Comprehensive software analysis:The software annotates mutations based on the WHO's "Catalogue of Mutations in Mycobacterium tuberculosis Complex and Their Association with Drug Resistance," covering all of the 1200+ Group1 and Group2 drug resistance-associated mutations (DRMs)and more than 7,000 DRMs in total. The amplified region spans 25.9 Kb and includes DRMs associated with 15 anti-tuberculosis drugs, relating to 52 genes or gene fragments. It supports MTBC identification, spoligotype typing, identification of 174 types of mycobacteria, and phylogenetic analysis.
Rapid full process:With DNBSEQ-G99ARS sequencer, the whole process from nucleic acid to report can be completed as fast as 24h. With DNBSEQ-E25 sequencer, the whole process from nucleic acid to report can be completed as fast as 30h;

Full workflow ofATOPlex Mycobacterium tuberculosis Targeted Sequencing Package
MTB-Explorer software report
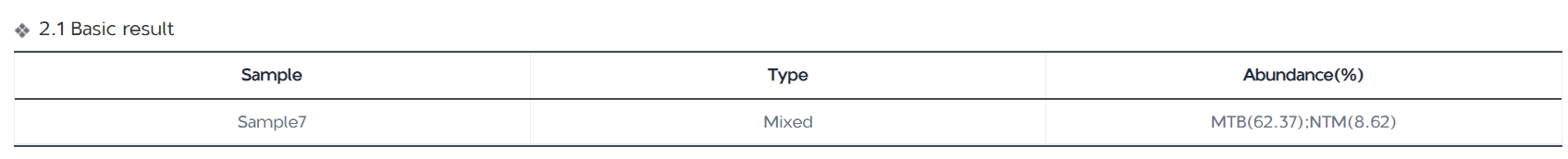
MTB-Explorer can generate summary reports for all samples analyzed in the same batch. Users can click on the corresponding links to access individual sample reports. The basic information page shows the mycobacterial identification results of the samples, types of mycobacterial infections, and the abundance ratio of mycobacteria in the samples (% of MTB reads/NTM reads).
The numbers of DRMs associated with 15anti-tuberculosis drugs detected in each sample will be visually displayed by a colorful circular graph.
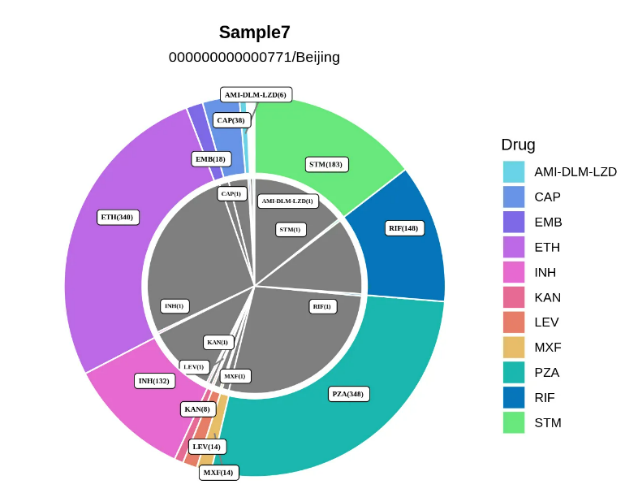
Remarks: The title indicates the name and pedigree of the sample, the colored outer circle indicates the number of variants associated with antituberculosis drug resistance included by WHO, and the gray inner circle indicates the corresponding number of resistance gene variants detected in the sample.
Drug resistance report result overview
The Mutation Detection Report page displays the number of all mutations detected in the samples, the number of resistance-associated mutations ( for group 1 and 2 variants in the WHO Catalogue of Resistance Variants); the number of indeterminate significance mutations and resistance-unassociated mutations detected (for group 3, 4, and 5 variants in the WHO Catalogue of Resistance Variants); and the number of other mutations detected (for those variants that have not been included in the WHO Catalogue). The report table also displays the mutation frequency, mutant base sequencing depth, and resistance confidence level annotations for each variant site; the IGV map provides a visual window into the mutant base sequencing depth at the corresponding position on the genome for each variant.
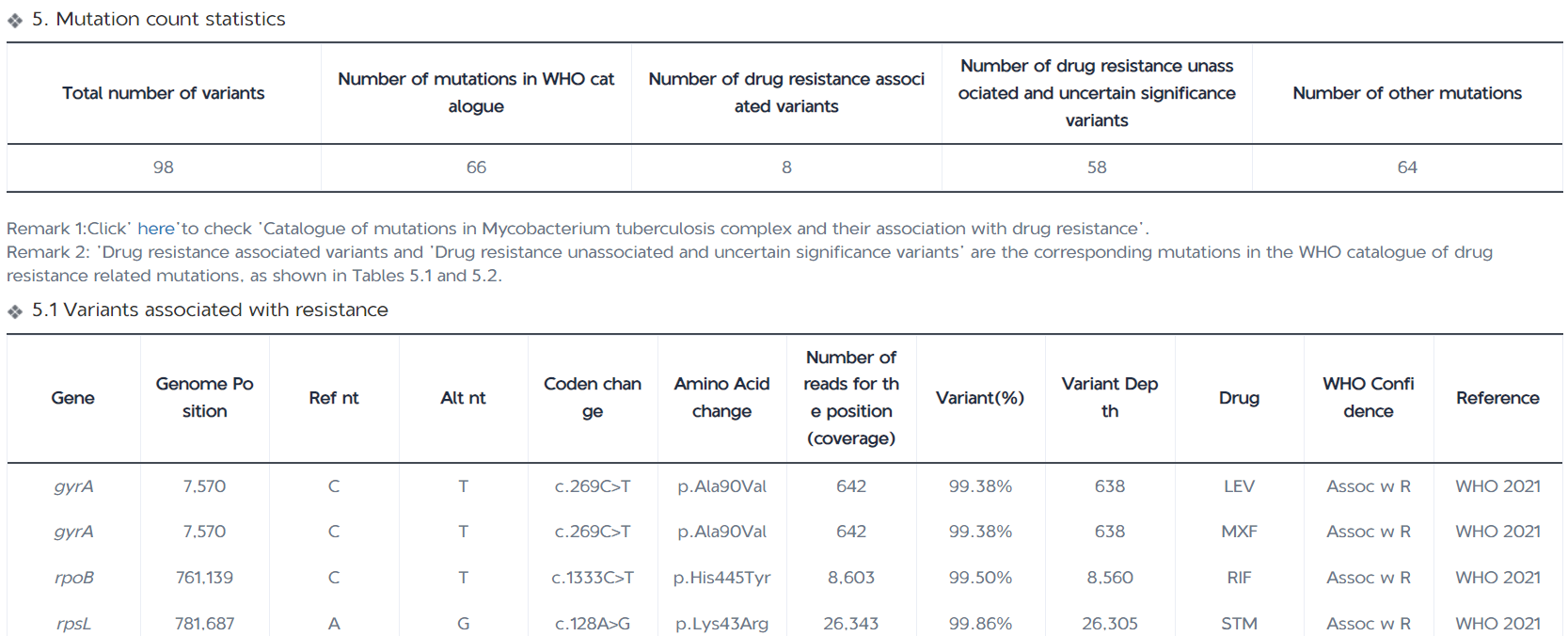
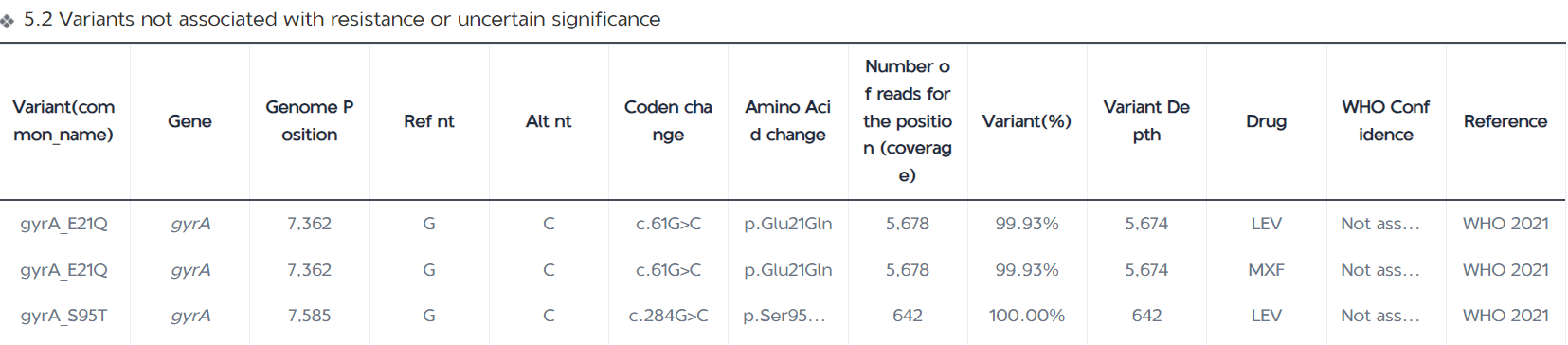
Statistics on the number and type of mutations
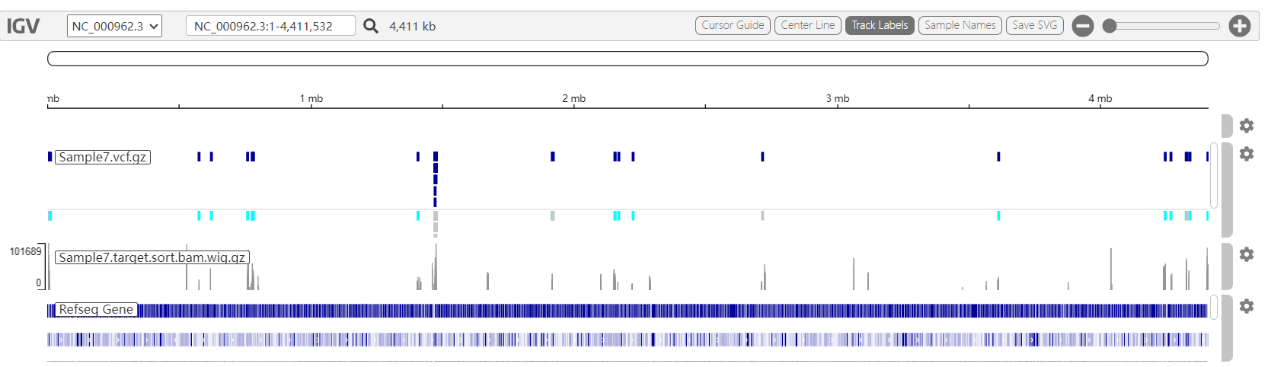
IGV Chart
The MTB-Explorer software summary report enables automatic preparation of evolutionary trees and SNP distance analysis heatmaps for all samples analyzed in the same batch.

Evolutionary tree analysis diagram
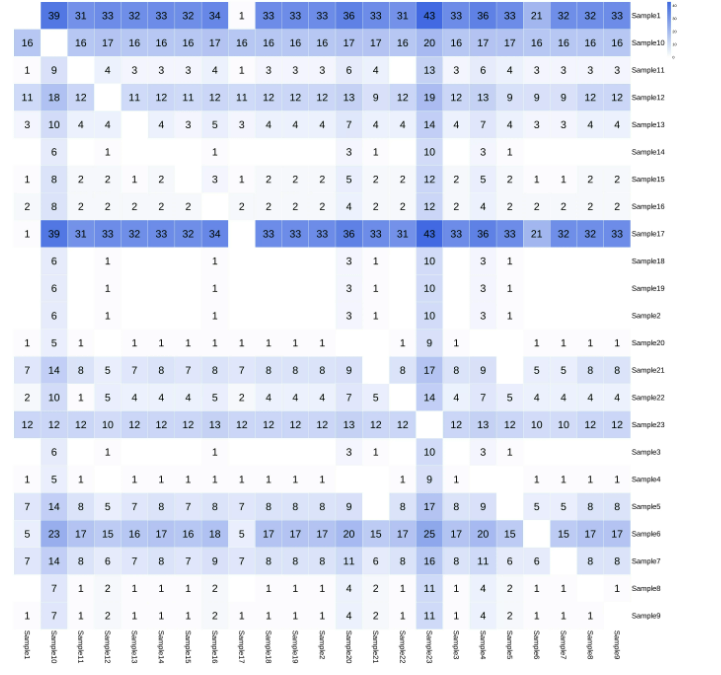
SNP distance analysis heat map
MGI has developed both HIV-1 drug resistance sequencing package and the ATOPlex Mycobacterium tuberculosis sequencing package. With their excellent data output quality and powerful software capabilities, these packages provide powerful tools to support pathogen mutation monitoring and drug resistance analysis.
In the field of public health, MGI has developed a diverse range of full-process combination products for various application scenarios, including metagenomic sequencing, targeted sequencing and whole genome sequencing of pathogenic microorganisms. Moving forward, adhering to the vision of "Leading Life Science Innovation", MGI will continue to innovate and provide more automatic, convenient and efficient core tools for pathogenic microorganism identification, drug resistance analysis and traceability. The commitment aims to contribute to the cause of public health prevention and control.
References:
1. Azevedo-Pereira J M, Pires D, Calado M, et al. HIV/Mtb co-infection: from the amplification of disease pathogenesis to an "emerging syndemic"[J]. Microorganisms, 2023, 11(4): 853.
Recommended ordering information for HIV-1 Drug Resistance Sequencing Products Package:
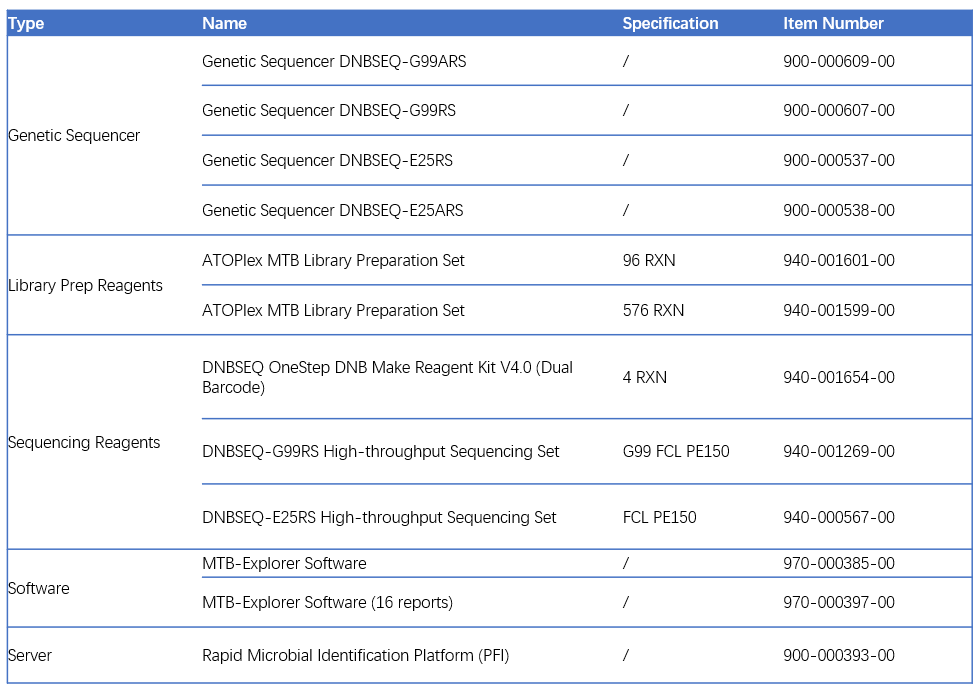
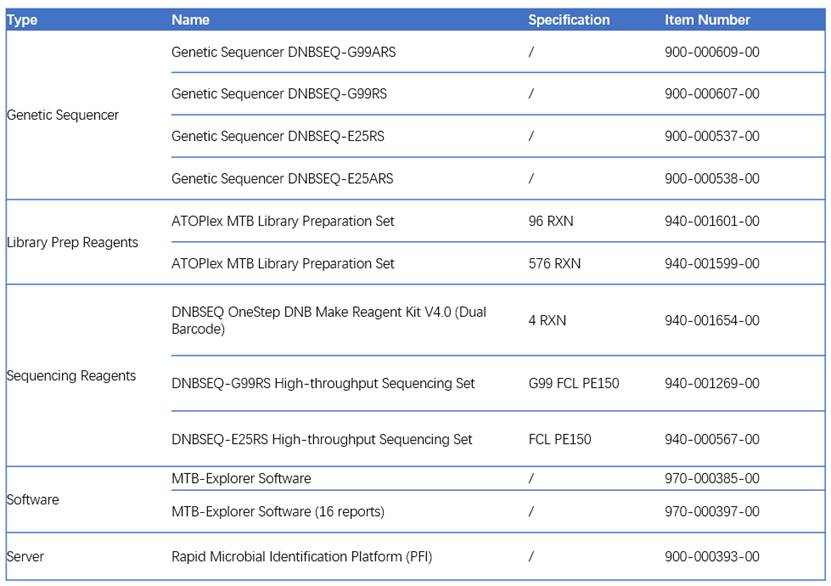
Recommended ordering information for ATOPlex Mycobacterium Tuberculosis Targeted Sequencing Package :
Datasheet-HIV-1 Drug Resistance Sequencing Products Package download

Brochure-ATOPlex Mycobacterium Tuberculosis Targeted Sequencing Package download




 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













