The human microbiome is a dynamic and complex microbial ecosystem, composed of trillions of host-specific microorganisms, including bacteria, viruses, fungi, and more. Human gut bacteria dominate this ecosystem, accounting for over 99% of the total gut microbial composition. An imbalance of gut microbiota can lead to various diseases, such as gastrointestinal disorders, metabolic diseases, immune and psychiatric disorders. Maintaining the balance of the human
microbiome is crucial for human health.
On July 23, MGI Tech launched Human Microbiome Metagenomics Sequencing Package, now is open for pre-order. This product package covers the entire process of human microbiome metagenomic sequencing, including sample collection, extraction, library preparation, sequencing, and analysis. It offers high automation and flexible throughput. Validated with numerous biological samples, it is an ideal tool for human microbiome research.
Highlights
Highly automated throughout the entire process, effectively reducing costs
To address the challenge of automated pretreatment of stool samples, MGI has developed an integrated stool collection kit, the MGIEasy Stool Collection Kit (GSC02), in combination with the MGISTP-7000 sample transfer processing system. This combination enables high-throughput and highly automated sample pretreatment. Using the MGISP-NE384, MGISP-960, and the MGISP-Smart8, the automation of nucleic acid extraction and library preparation is realized, saving both time and effort. For library preparation, MGI optimized the assay, reducing the reagents by half and effectively cutting the cost.
Flexible and efficient one-stop solution
MGI offer a flexible product package, allowing them to choose different throughput automated instruments and sequencers based on sample volume, ranging from 15,000 to 450,000 samples per year. Combined with the Platform of Microorganisms Fast Identification (PFI), it can identify and analyze microorganisms quickly and accurately.
Excellent performance verified by a large number of individuals’ stool samples
Over 130,000 stool samples have been validated on DNBSEQ platforms through the Million Microbiomes from Humans Project (MMHP). The sequencing results exhibit excellent intra-platform repeatability (relative abundance Spearman's correlation coefficient: genes >0.91, species >0.97) and high consistency with the results from other vendors' sequencer (gene richness Spearman's correlation coefficient >0.99).

Figure 1. MGI Human microbiome metagenomics sequencing package
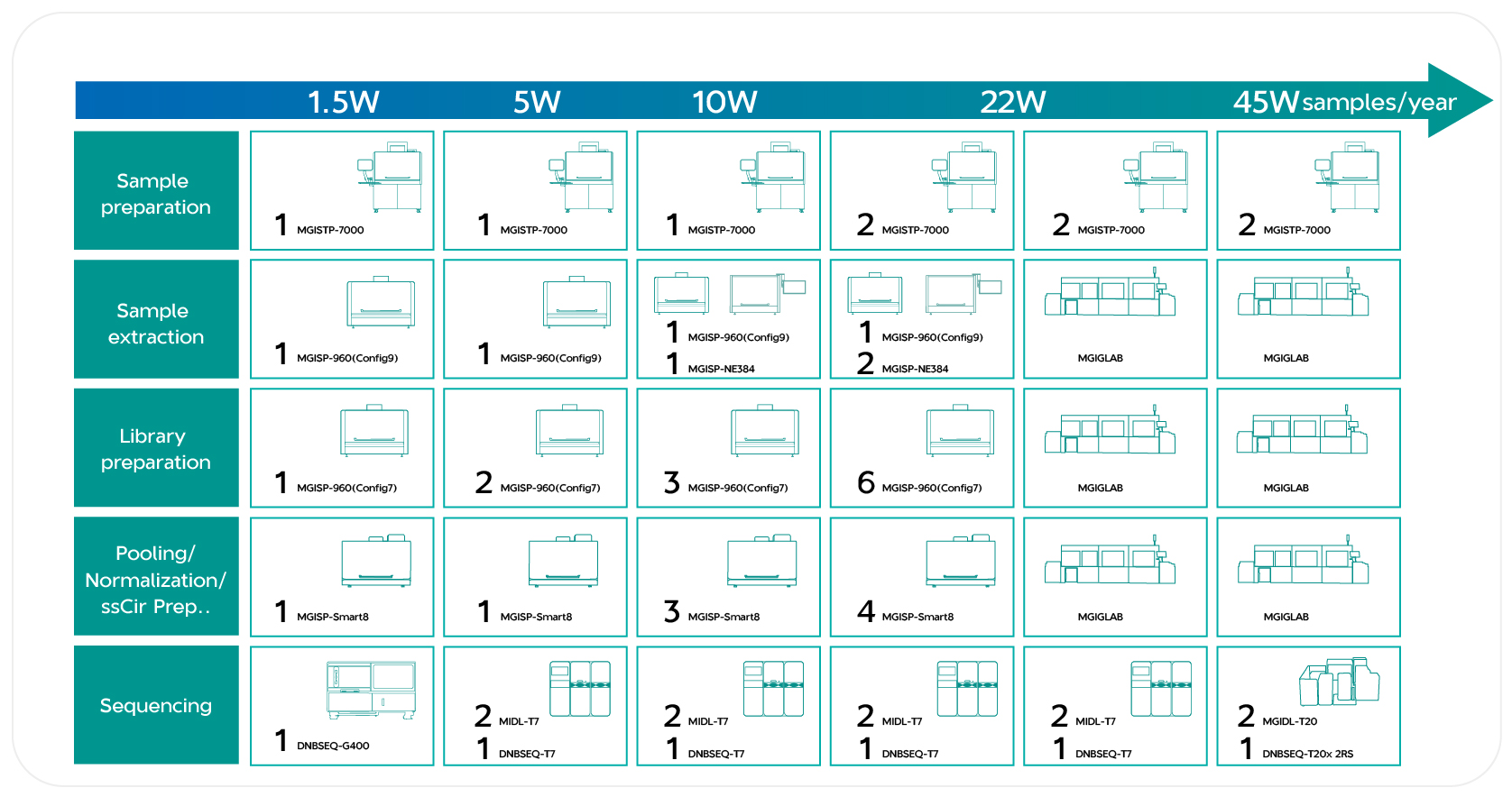
Figure 2. Combination product with different throughputs
Performance
1. The MGIEasy Stool Collection Kit combined with the MGISTP-7000 enables high-throughput automated sample pretreatment.
Using the MGIEasy Stool Collection Kit (GSC02-I-15), 73 stool samples were randomly collected, with the sample weight ranging from 0.9 to 2.1 g (average sampling weight of 1.67 g, Figure 3 A). After thorough mixing, MGISTP-7000 automatically decap the sample tubes and transfer 1 mL of
the sample to 96-well plate. The success rate of automated sample transferring for the 76 samples (including 3 blank controls) is 100%, with CV of pipetting volume < 15%, meeting the requirements for automated sample pretreatment (Figure 3 B).

Figure 3. Results of automated sample pretreatment using the MGIEasy
Stool Collection Kit (GSC02-1-15) combined with MGISTP-7000.
2. MGI' automated system can achieved precision extraction and library preparation
Samples were extracted using the MGIEasy Stool Genome DNA (meta) Extraction Kit II with MGISP-960. 95.9% of the extracted gDNA concentration more than 8 ng/μL, and all the samples met the risky library preparation standard (> 2 ng/μL). The purity, with A260/A280 ratio of 1.7 to 2, meet the requirements for metagenomic library preparation (Figure 4 A, B, C). Using the MGIEasy Fast FS DNA Library Preparation Kit Set V2.0 and MGISP-960 with both standard DNA input (200 ng) and low DNA input (10 ng - 50 ng), over 95% of the libraries achieved a concentration higher than 10 ng/μL, meeting the quality control standards (Figure 4 D).
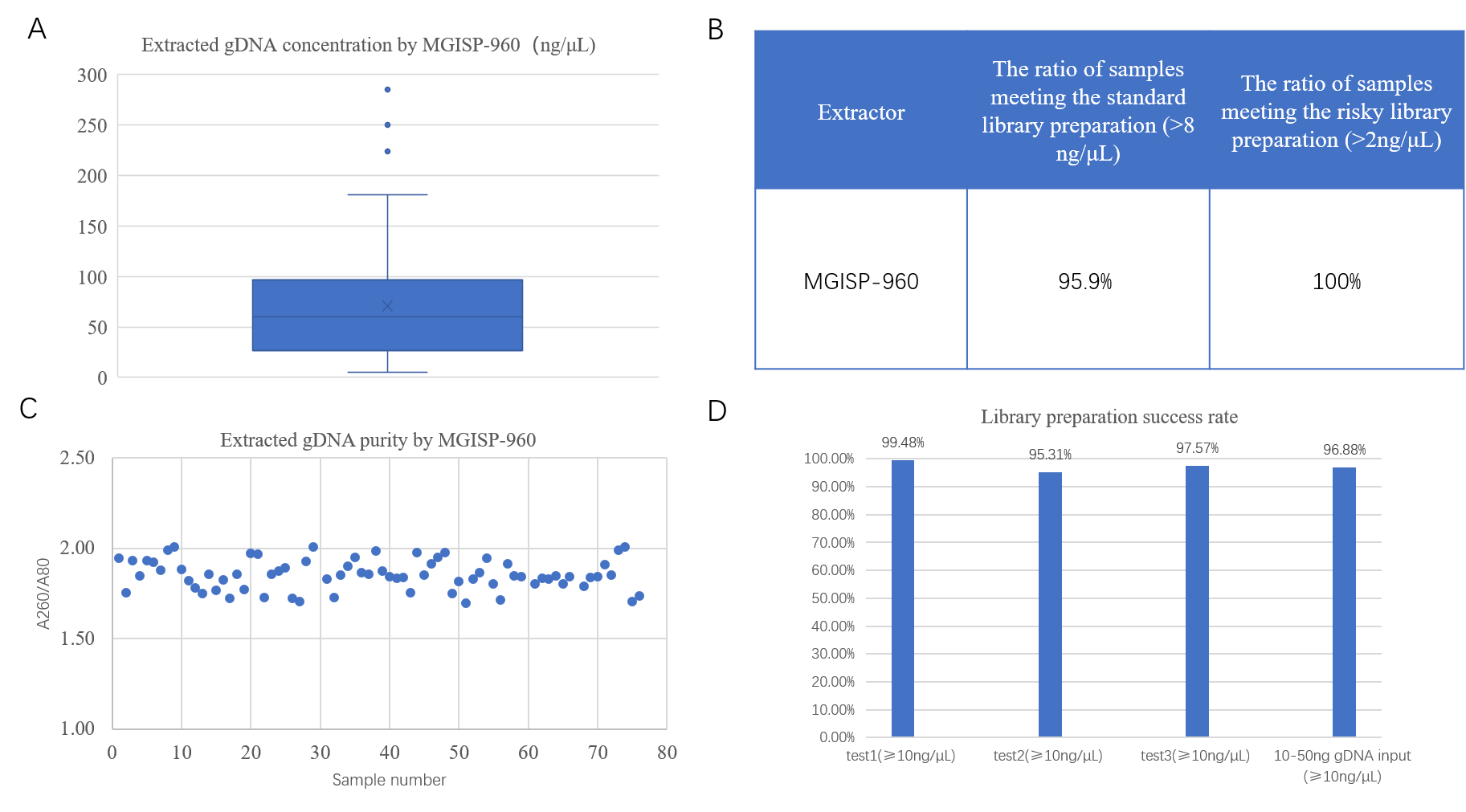
Figure 4. The results of automated extraction and library preparation utilizing the MGISP-960
3. The perfect combination of automated Systems, DNBSEQ Platform and PFI enables accurate analysis of species abundance
Mock samples were sequenced on the DNBSEQ-G400 and DNBSEQ-T7 platforms, with microbial identification and abundance analysis performed using PFI software. The results showed a 100% microbial detection rate across various gDNA inputs (10, 25, 50, 200 ng) and sequencers (DNBSEQ-G400, DNBSEQ-T7) (Figure 5A). Comparative analysis revealed that bacterial abundance was highly consistent under different conditions, with R² values all above 0.99 (Figure 5B).
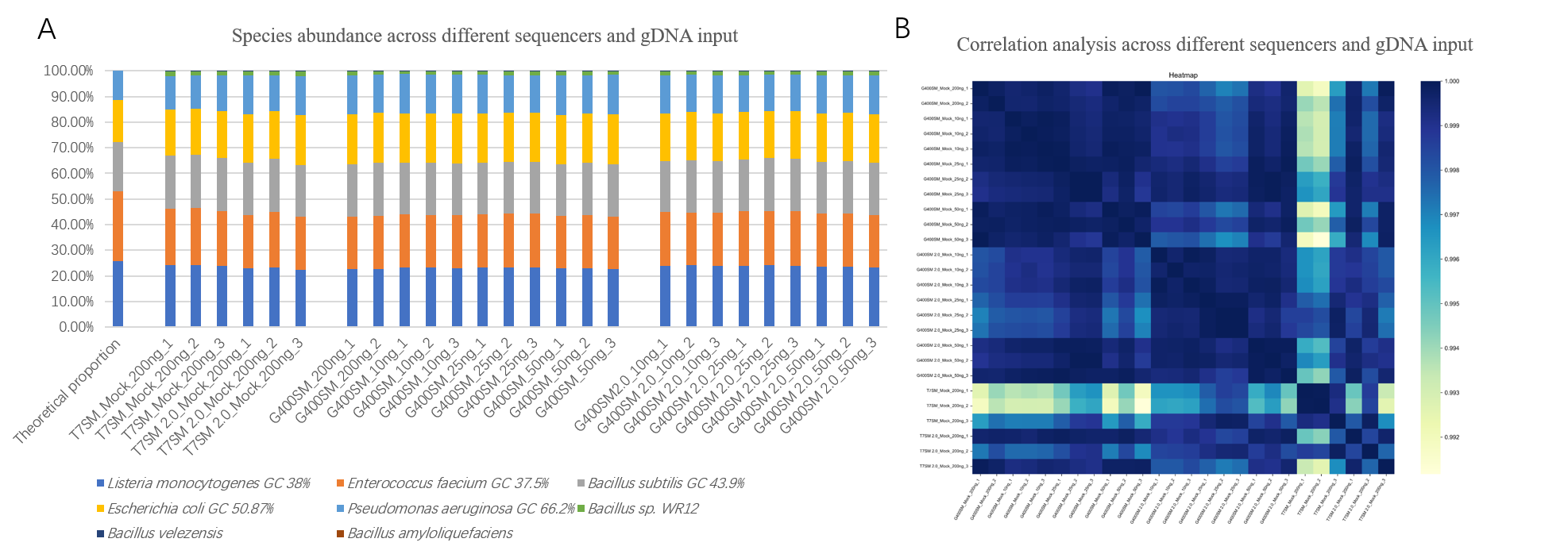
Figure 5. The relative abundance of different bacteria
detected by DNBSEQ-G400 and DNBSEQ-T7.
SM 2.0 represents StandardMPS 2.0, an upgraded sequencing reagent
that can achieve sequencing quality up to Q40.
Compared to traditional techniques, metagenomics is increasingly welcomed by the scientific community for its speed and accuracy. The MMHP consortium and the MOHA Initiative are committed to making metagenomics accessible to all by transferring technology to reference
laboratories. MGI's one-stop Human microbiome metagenomics sequencing package, with its
standardized, professional, and automated processes, ensures high quality and high precision of sequencing data, setting a new benchmark for human microbiome research and is expected to greatly promote innovation in translational medicine.
Adhering to the vision of "Leading Life Science Innovation ", MGI continues to forge ahead on the path of innovation. In the future, with the mission of being a creator of core tools in life science, MGI will persist in deepening and expanding in the field of life science, strengthening innovation, bringing more innovative technologies and products to users worldwide, and empowering the development of the life science industry.
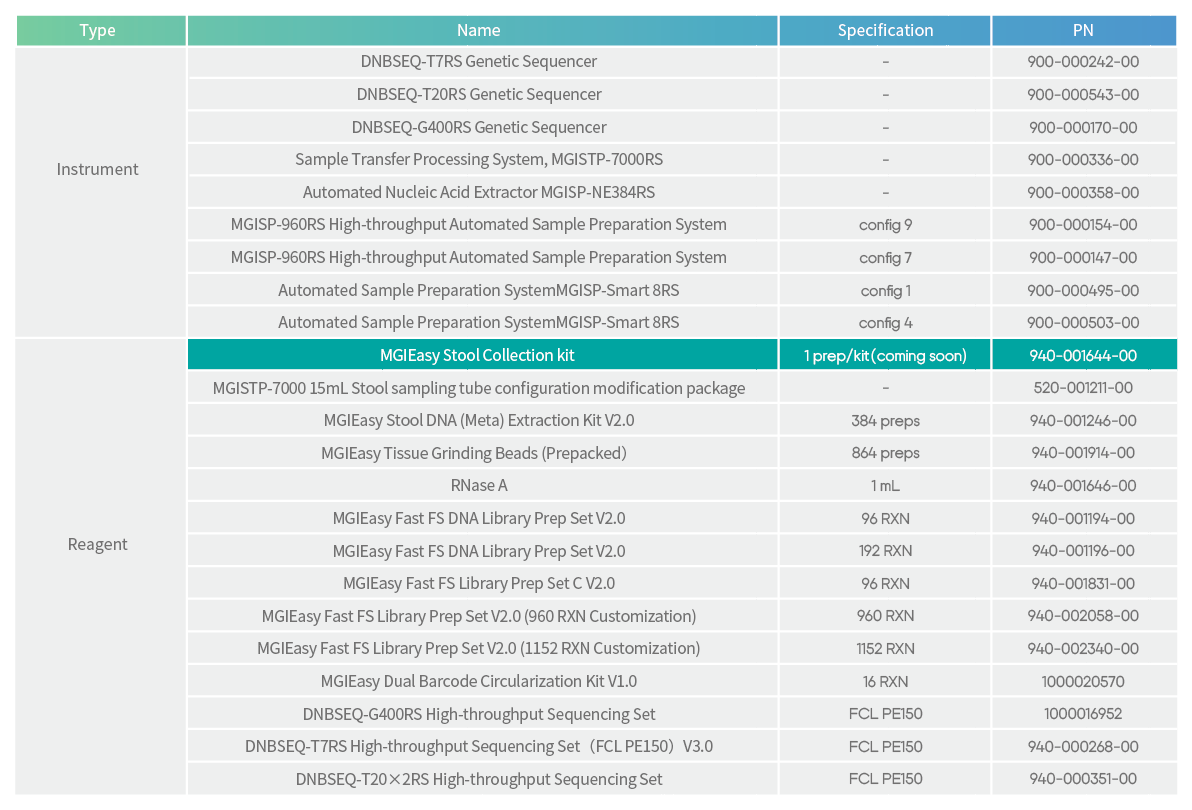
Ordering information
Reference:
1. Chu J, Feng S, Guo C, et al. Immunological mechanisms of inflammatory diseases caused by gut
microbiota dysbiosis: A review[J]. Biomedicine & Pharmacotherapy, 2023, 164: 114985.
2. Altveş S, Yildiz H K, Vural H C. Interaction of the microbiota with the human body in health and diseases[J]. Bioscience of microbiota, food and health, 2020, 39(2): 23-32.
3. Kho Z Y, Lal S K. The human gut microbiome–a potential controller of wellness and disease[J]. Frontiers in microbiology, 2018, 9: 1835.
4. Meslier V, Quinquis B, Da Silva K, et al. Benchmarking second and third-generation sequencing
platforms for microbial metagenomics[J]. Scientific Data, 2022, 9(1): 694.



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













