Shenzhen, China, 20 November 2024 – MGI Tech Co., Ltd. (“MGI”), a company committed to building core tools and technologies that drive innovation in life science, lately launched its ATOPlex DENV1-4 Targeted Sequencing Package, a comprehensive solution for the rapid and accurate detection of dengue virus (DENV). This innovative package offers a streamlined workflow encompassing nucleic acid extraction, library preparation, DNBSEQ™ sequencing, and data analysis, delivering results in as little as 12 hours to aid the rapid detection of dengue fever outbreaks.
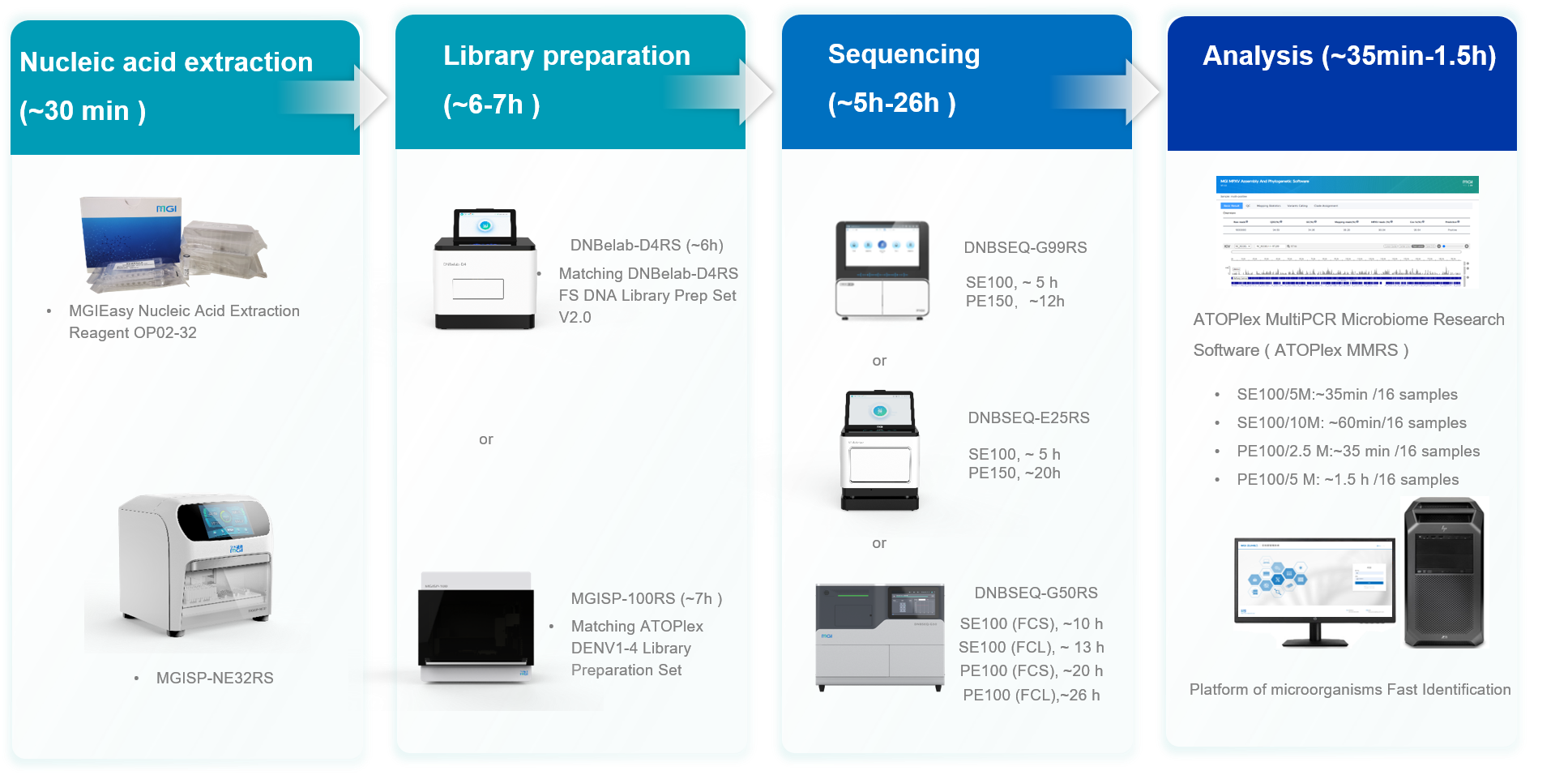
Figure 1. MGI ATOPlex DENV1-4 Targeted Sequencing Package
Dengue fever, a prevalent mosquito-borne disease in tropical and subtropical regions caused by dengue virus, poses a significant public health threat. As prevalence remains high in urban and semi-urban areas worldwide, MGI’s new ATOPlex DENV1-4 Targeted Sequencing Package provides a convenient and powerful tool for timely and precise detection of DENV, crucial for effective outbreak control and management.
The package includes the ATOPlex DENV1-4 Library Preparation Set, which leverages MGI's proprietary ATOPlex multiplex PCR technology, enabling easy operation and requiring minimal sample input. It also offers flexible sequencing solutions with SE100 for faster turnaround time and PE100 for cost-effectiveness, catering to diverse user needs.
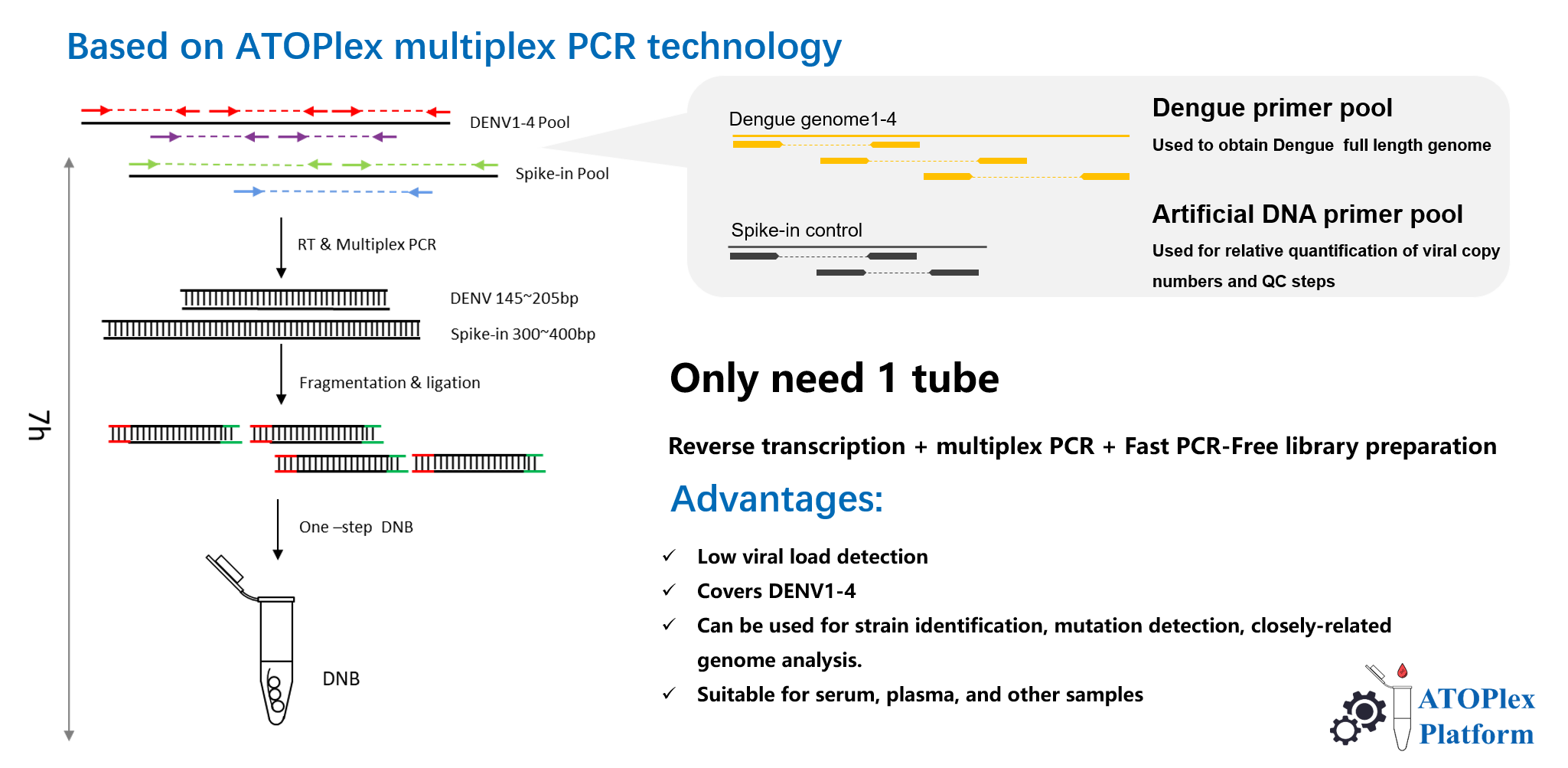
Figure 2. Library preparation principle
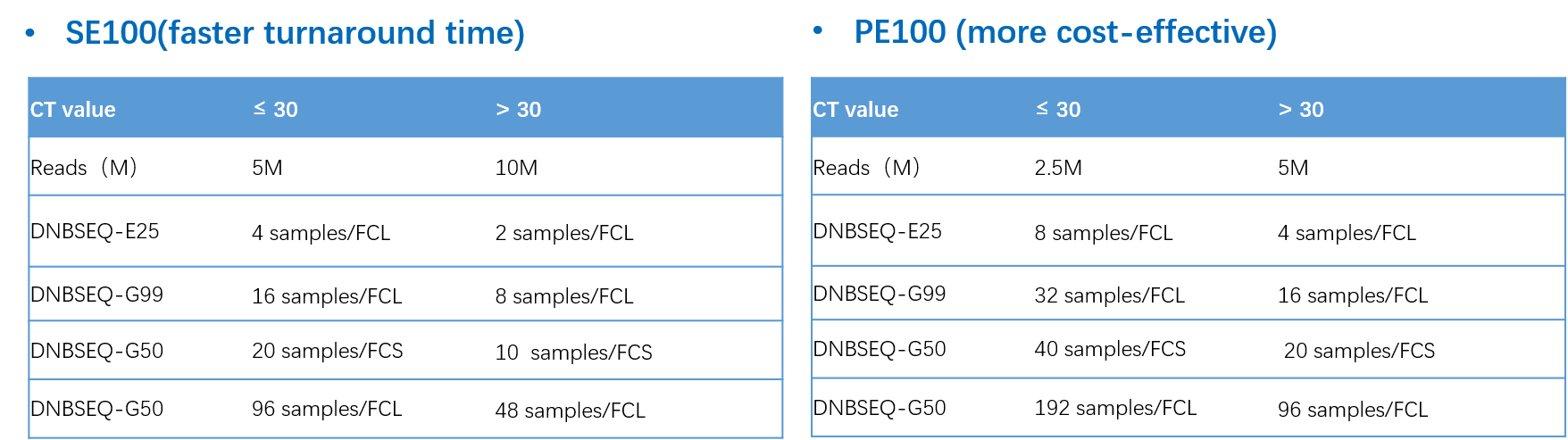
Figure 3. Different solutions
Additionally, the ATOPlex MultiPCR Microbiome Research Software (ATOPlex MMRS) simplifies data interpretation and provides accurate typing results based on built-in reference genome sequences and primer information for several target microorganisms, including dengue virus, coronavirus, mpox virus, respiratory syncytial virus, and any custom input species.
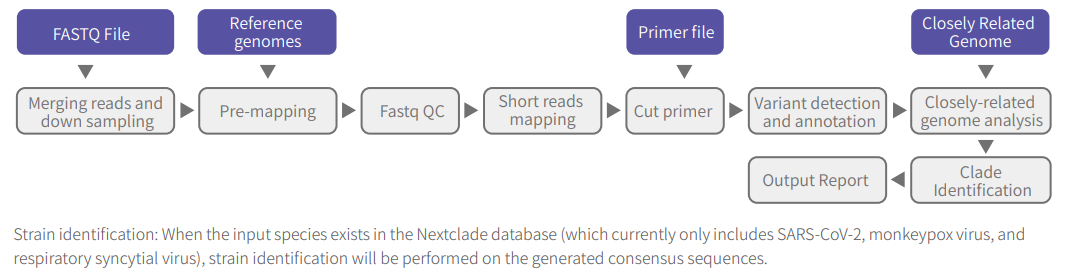
Figure 4. ATOPlex MMRS analysis workflow
Performance validation demonstrates exceptional performance metrics, with a 100% success rate in library preparation and superior yields compared to manual methods. All DNB concentrations met sequencing quality thresholds. At the same time, it showcased high sequencing quality across different DNBSEQ platforms with more than 95% of bases scoring Q30. The average quality control values at 1x, 10x, and 100x sequencing coverage all surpassed 99% on all tested platforms. Moreover, the typing results of dengue fever samples were consistent with known results, validating the package's reliability.
With an input of 200 ng, different RNA samples (DF and M) were prepared, yielding a library output of ≥20 ng in all cases, with a 100% success rate in library preparation. Additionally, the library yields produced by MGISP-100RS were generally higher than those obtained manually (Figure 5). Furthermore, the DNB concentrations consistently measured ≥8 ng/μL (Table 1), successfully meeting the requirements for further sequencing.
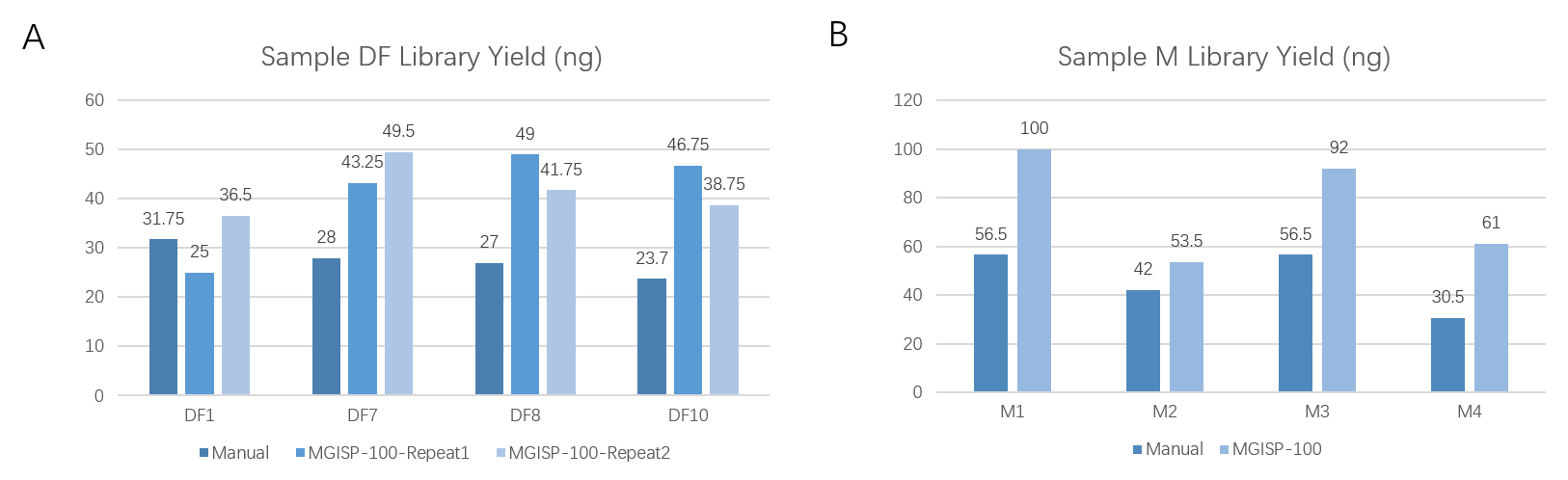
Figure 5. Library yields from manual and MGISP-100RS for DF and M samples

Table 1. DNB making results
Different DNBs were loaded onto different sequencing platforms: DNBSEQ-G99ARS, DNBSEQ-E25RS, and DNBSEQ-G50RS (SE100+10+10), the Total Reads all met the requirements of more than 80M, 25M, and 100M, respectively, and the Q30 of each flow cell was greater than 95% (Table 2). At the same time, the average values of 1×Coverage(%), 10×Coverage(%), and 100×Coverage(%) of the three sequencing platforms were all greater than 99 % (Figure 6 ), and the sequencing results produced by MGISP-100 were comparable to those obtained manually (Figure 6C , D) .
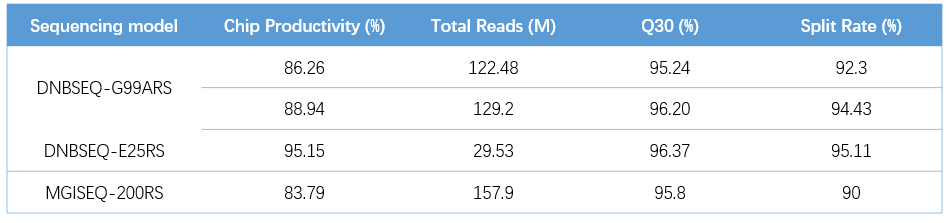
Table 2. QC metrics of different sequencing platforms
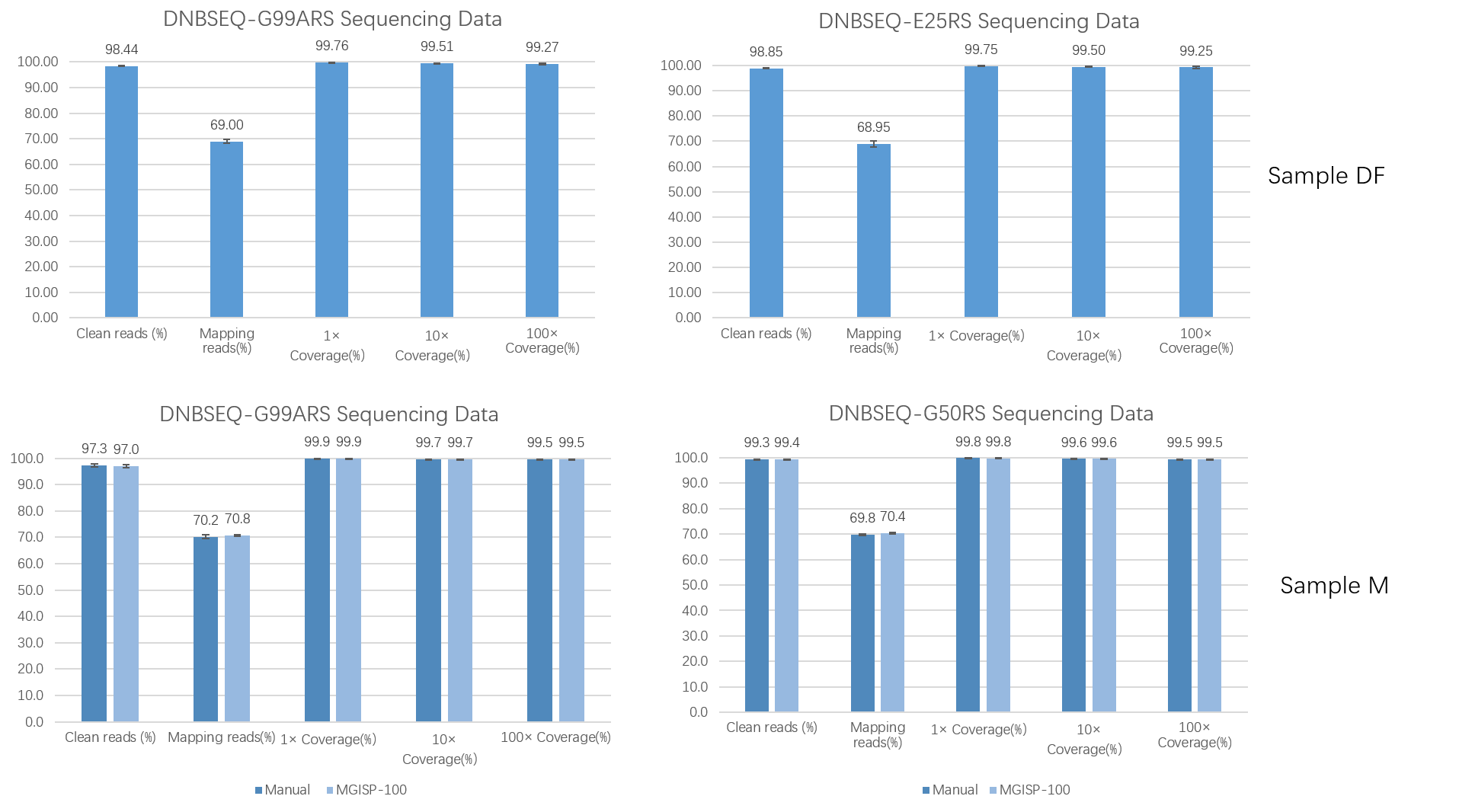
Figure 6. QC metrics of different sequencing platforms.
(A, B) DF samples were prepared by MGISP-100
and the sequencing results were obtained on DNBSEQ-G99ARS / DNBSEQ-E25 ;
(C, D) M samples were prepared by manual/
MGISP-100 combined with DNBSEQ-G99ARS / DNBSEQ-G50RS for sequencing
Further clade identification analysis revealed that the typing results of DF samples were consistent with the known results, DF1 was DENV2 , DF7 , DF8 , and DF10 were DENV1. All M samples were detected to be DENV1(Table 3).
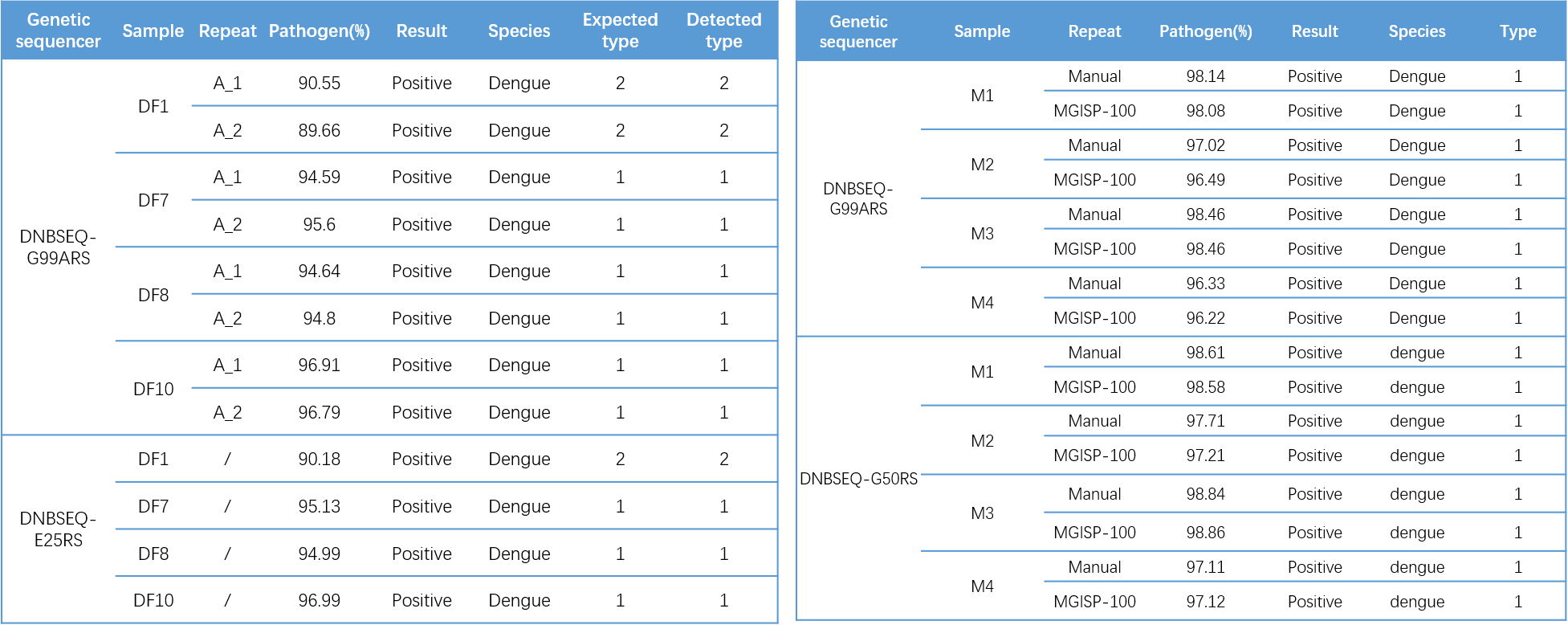
Table 3. Typing results of DF (left) and M (right) samples
A pilot program using the ATOPlex DENV1-4 targeted sequencing process has been deployed since late 2023 as part of a dengue fever traceability study with multiple Centers for Disease Control and Prevention in Shantou, Jieyang, and Chaozhu in China. Sequencing results from the three cities indicated that dengue fever cases in eastern Guangdong may have originated from different strains, demonstrating the package’s real-world applicability.
Committed to leading life science innovation and providing cutting-edge tools for public health, MGI’s ATOPlex DENV1-4 Targeted Sequencing Package empowers researchers and public health officials with an accurate and efficient solution for important dengue fever detection, monitoring, prevention, and research, contributing to improved outbreak control and effectively informing public health initiatives for better health outcomes.
The ATOPlex MultiPCR Microbiome Research Software Analysis Package (16 reports) is now available for free trial. For details, please scan the QR code.

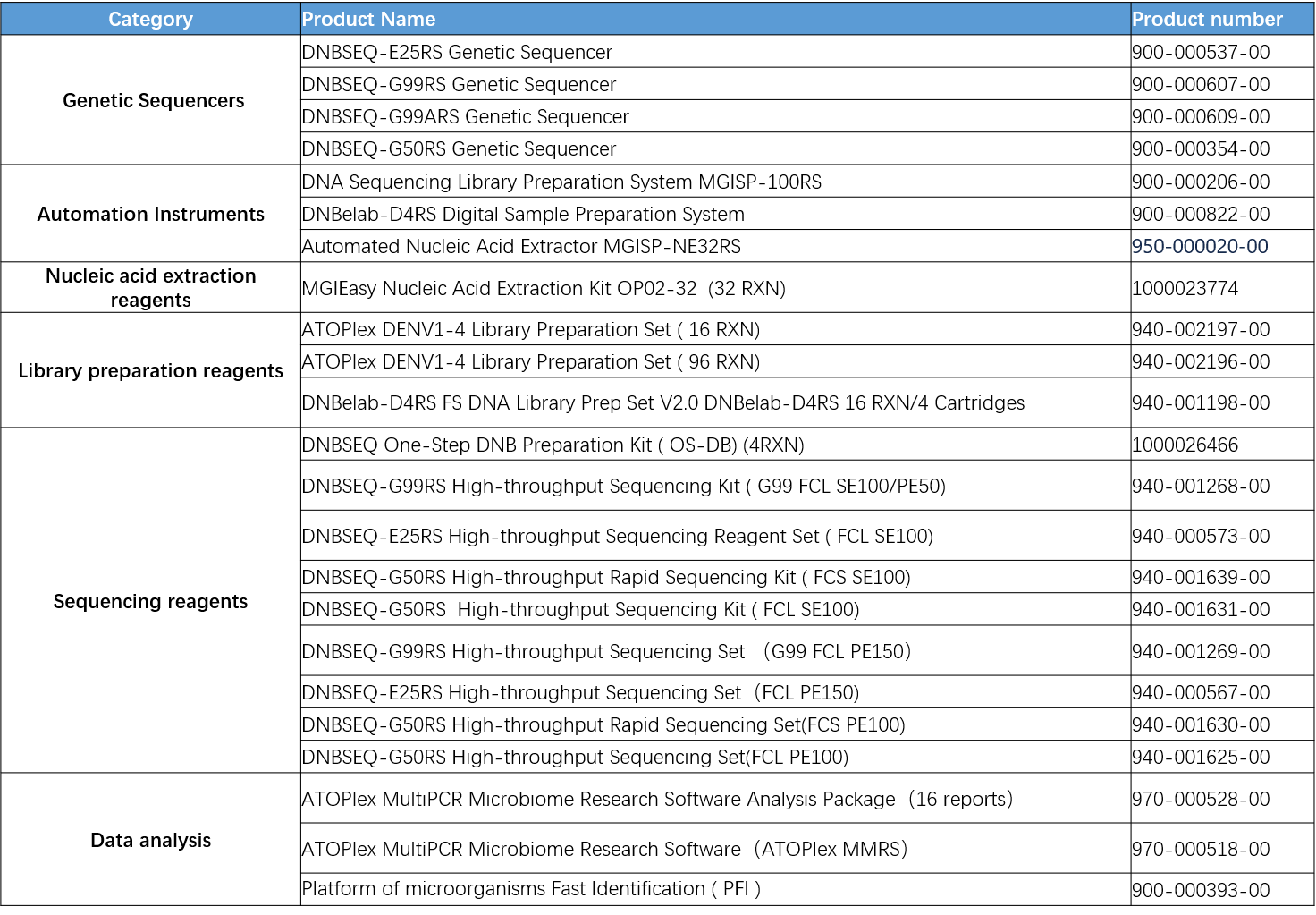
Recommended ordering information:



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













