Shenzhen, China, 28 February 2025 – MGI Tech Co., Ltd. (“MGI”), a company committed to building core tools and technologies that drive innovation in life science, has launched the latest Microbiome Metabarcoding Sequencing Package (“MMSP”), specifically designed to support scientists in microbiome research. This cutting-edge package utilizes the DNBSEQ-G99 and DNBSEQ-E25 sequencing platforms and is now available for global orders.
Microbiomics explores the behavior, interaction, and function of microbial communities in specific environments. Microbiome sequencing represents a groundbreaking technology that allows researchers to delve into and understand the complexity of microbial communities in the human body and the environment, revealing the diversity, abundance, and function of microorganisms. It shed a light on understanding how microorganisms affect human health, ecological environment, industrial and agricultural production. 16S/ITS rDNA sequencing provides a rapid and economical method for microbial classification at the genus - species level. It is a widely used technique for microbial community analysis.
Applications of microbiome in human body, environment, and industrial and agricultural fields[1]

Workflow Demonstration of MGI Microbiome Metabarcoding Sequencing Package
Key Features of MGI MMSP:
l Versatile Applications: Designed around the popular variable regions of rDNA, MGI MMSP is compatible with a wide range of sample types to meet diverse demands across various applications, including human, industrial, agricultural, and environmental microbiome research.
l Comprehensive Solution: MGI MMSP integrates nucleic acid extraction, library preparation, sequencing, and analysis into a single, streamlined workflow, simplifying the research process for scientists..
l Flexibility: The MGI MMSP can be operated on small or medium-sized sequencing platforms. With the DNBSEQ-G99, researchers can achieve ultra-fast PE300 sequencing. Additionally, the DNBSEQ-E25 allows for portable sequencing, offering ease of use in challenging environments—even on a bumpy boat.
Performance References
- Sample type 1: Zymo standard
The following two sets of data were obtained by sequencing and analyzing the Zymo standard with 16SV3V4 and 16SV4 to evaluate the species detection rate, species abundance accuracy, and repeatability. As shown in Figures 1 and 2, all expected species were detected, the measured abundance was close to the theoretical abundance, and the repeatability between and within batches was good.
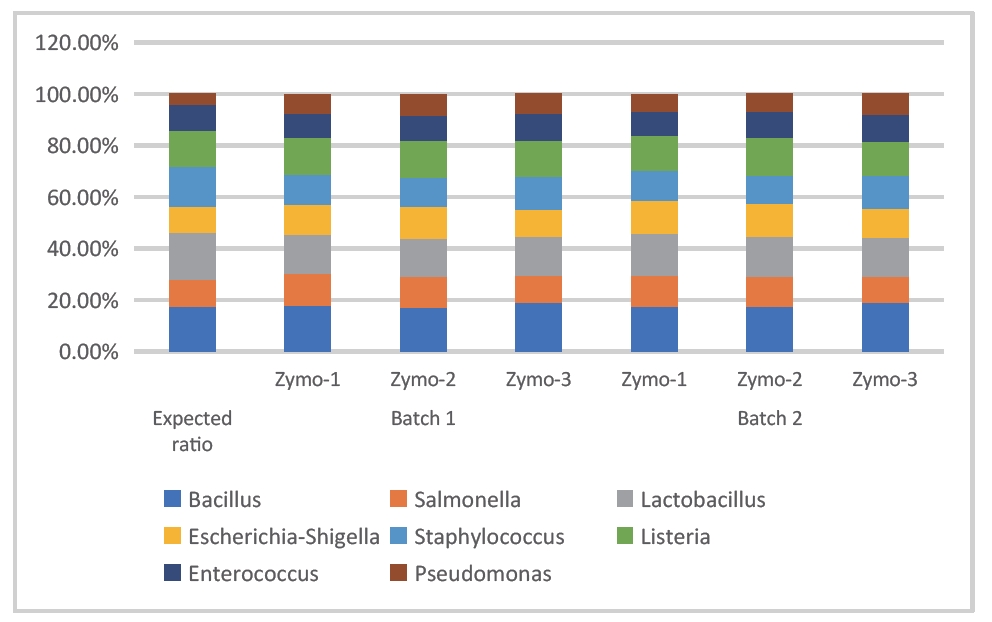
Figure 1: Stacked bar chart of bacterial abundance in Zymo standard (DNBSEQ - G99_16SV3V4)
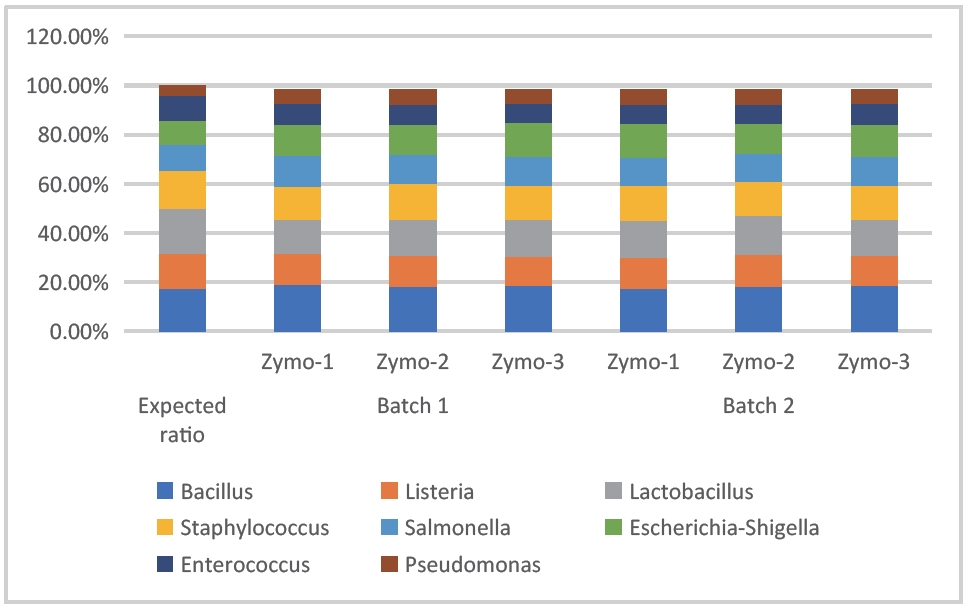
Figure 2: Stacked bar chart of bacterial abundance in Zymo standard (DNBSEQ - E25_16SV4)
- Sample type 2: Human feces
The following data were obtained by sequencing and analyzing human feces with 16SV3V4 and ITS2. As shown in Figures 3 and 4, Prevotella, Alloprevotella, Lachnospira, and Saccharomyces, Candida, Nigrospora, which are common bacterial and fungal communities in the gut, were identified respectively.
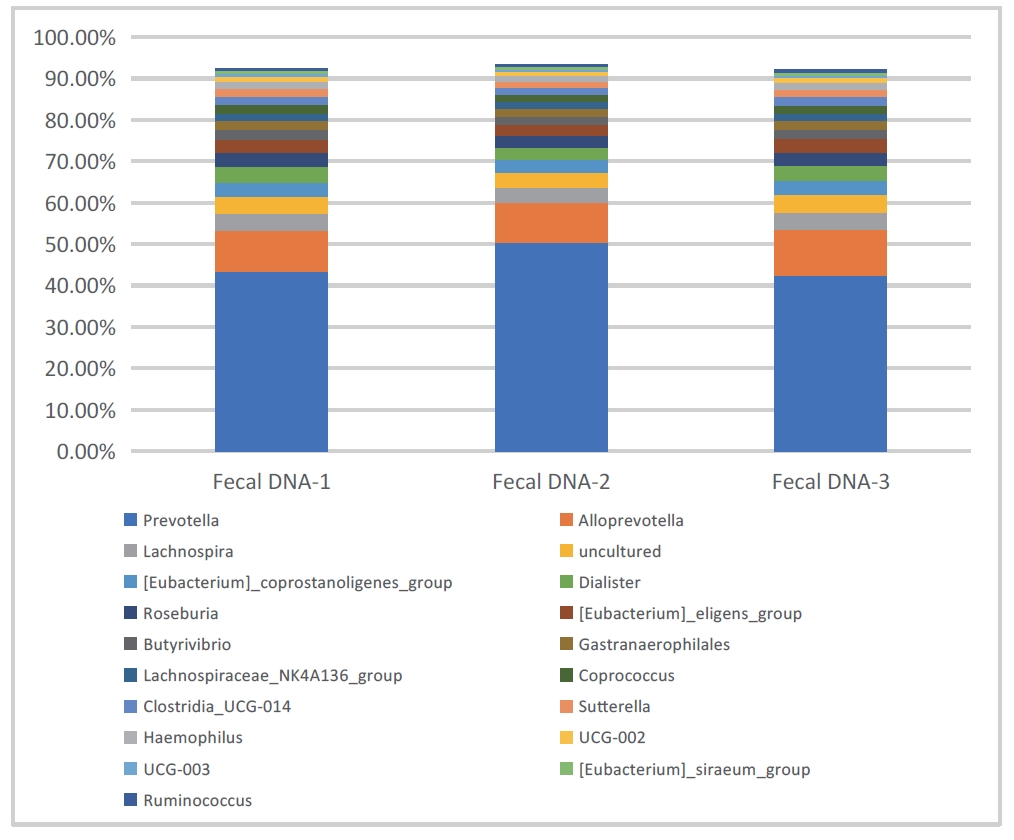
Figure 3: Stacked bar chart of bacterial abundance in fecal sample (DNBSEQ - G99_16SV3V4)
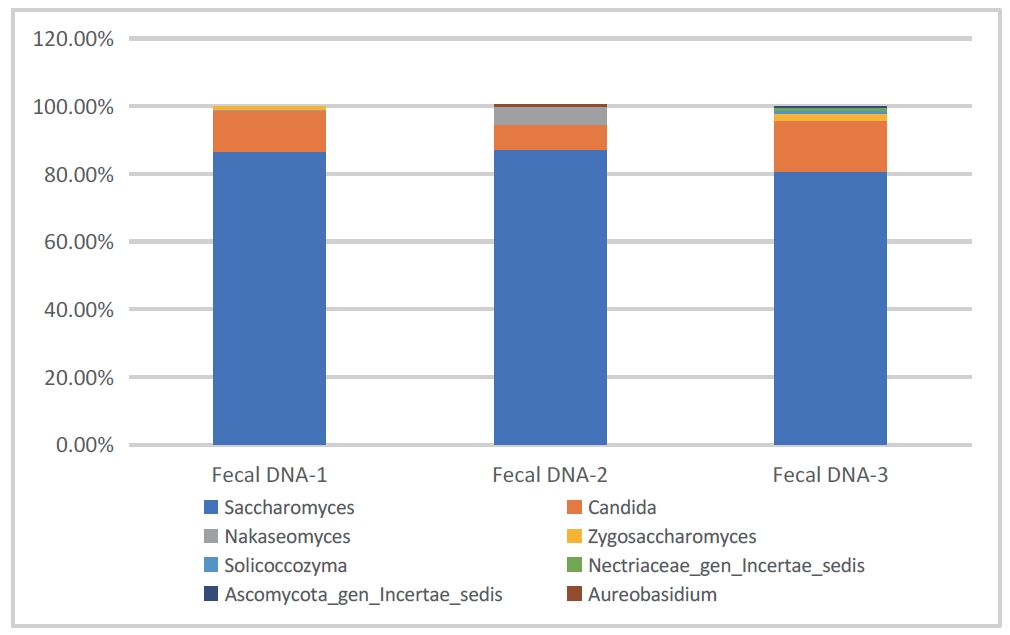
Figure 4: Stacked bar chart of bacterial abundance in fecal sample (DNBSEQ - G99_ITS2)
- Sample type 3: Fermented grain
The following data were obtained by sequencing and analyzing the fermented grain with 16SV4 and ITS1. As shown in Figures 5 and 6, Weissella, Lactobacillus, Oceanobacillus, and Pediococcus, which are common bacterial communities in the fermented grain, and Mucor, Paecilomyces, Aspergillus, which are common fungal communities, were identified respectively.

Figure 5: Stacked bar chart of bacterial abundance in fermented grain (DNBSEQ - E25_16SV4)
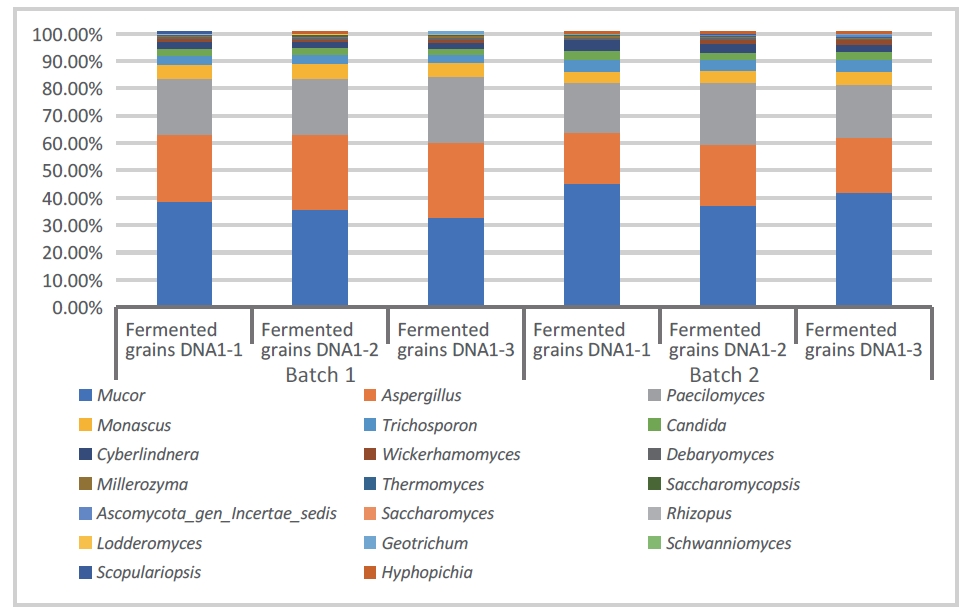
Figure 6: Stacked bar chart of bacterial abundance in fermented grain (DNBSEQ - G99_ITS1)
- Sample type 4: Leaf surface
The following data were obtained by sequencing and analyzing the leaf surface washing solution with ITS1. As shown in Figure 7, Cladosporium, Nigrospora, Curvularia, Elsinoe, Lachnum, which are common fungal communities causing leaf diseases, were identified.
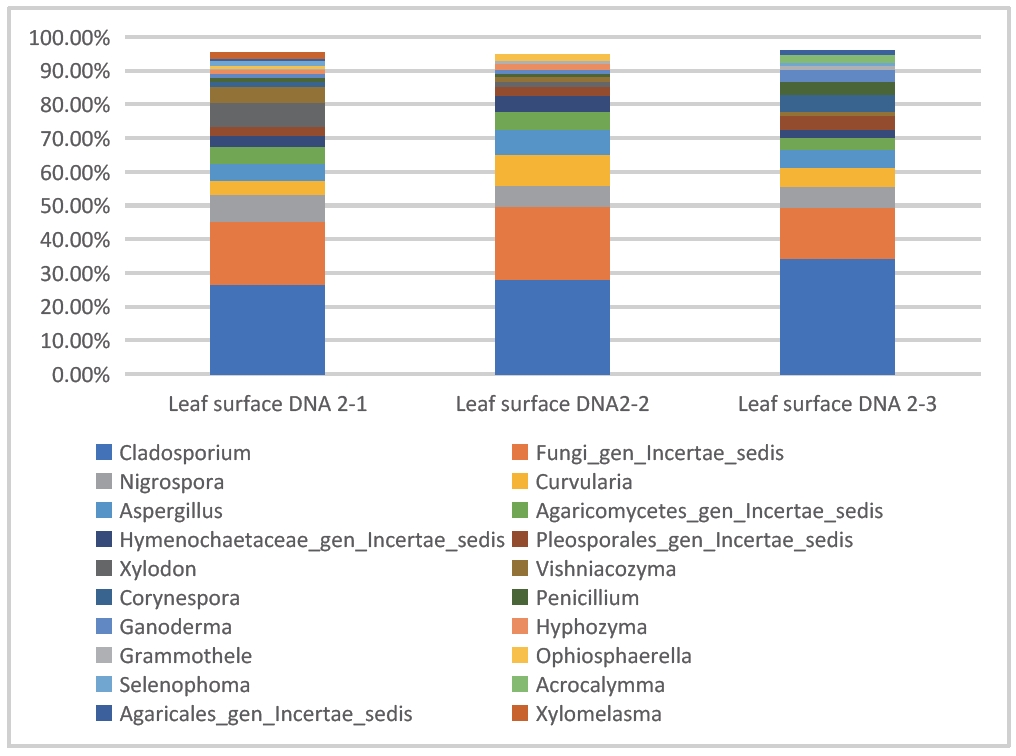
Figure 7: Stacked bar chart of bacterial abundance on leaf surface (DNBSEQ - G99_ITS1)
- Sample type 5: Deep sea sediment in the South China Sea
The following data were obtained by 16SV4 rDNA sequencing the deep sea sediment sample in the South China Sea using DNBSEQ - E25 on boat. As shown in Figure 8, uncultured bacteria and archaea, such as green curved bacteria, acid - rod bacteria, and deformed bacteria, were detected in the sample. Functional gene annotation was also performed, providing an important reference for deeply understanding of the biogeochemical mechanisms of important microbial groups in the deep sea.
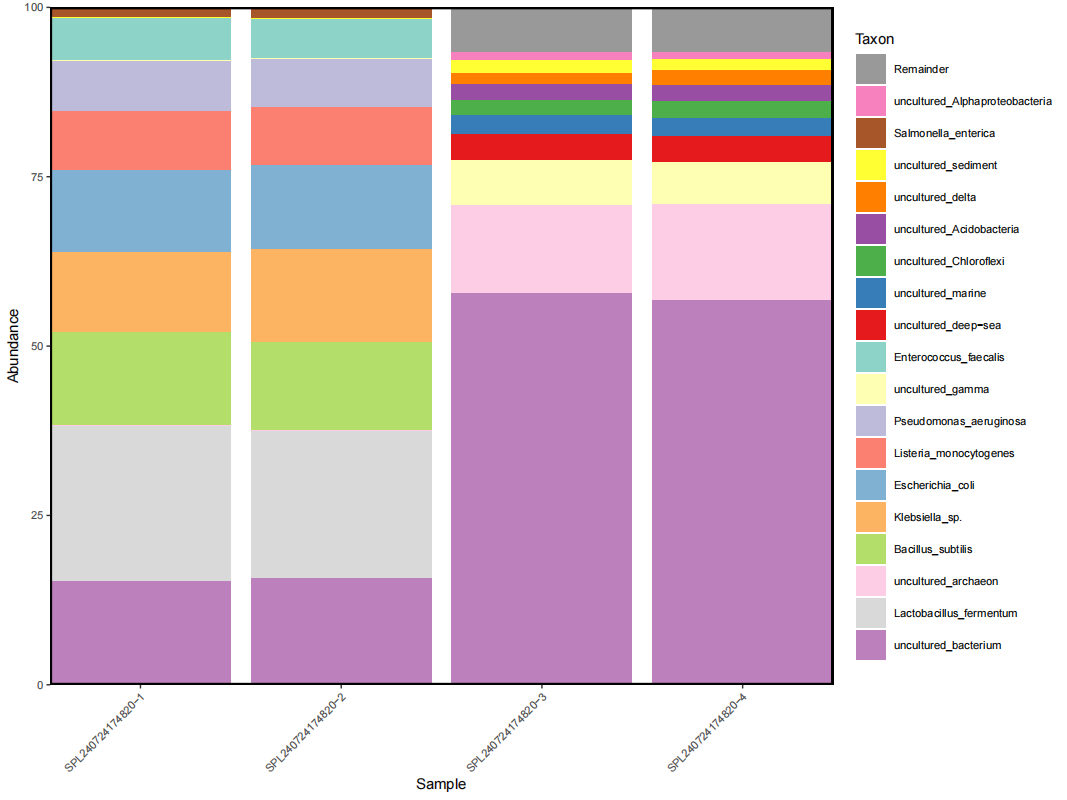
Figure 8: Stacked chart of species distribution
It has been verified that MGI Microbiome Metabarcoding Sequencing Package work for various sample types, covering a wide range of application scenarios in the field of microbiology, and has excellent detection performance and powerful analysis functions. It can be an ideal tool for microbiome research.
For more details and order information, please visit: https://en.mgi-tech.com//Uploads/Temp/file/20250226/67bee77b802b5.pdf



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













