Shenzhen, China, 21 May 2025 - MGI Tech Co., Ltd. (“MGI”), a company dedicated to developing core tools and technologies that drive innovation in life sciences, today announced the launch of the Microbial Isolates Whole-Genome Sequencing Package (for CycloneSEQ) and its global availability for ordering. The package aims to empower microbial genomics research with a comprehensive sequencing solution that offers a seamless operation experience with a 3-step-only library preparation design. Additionally, MGI also launched and announced the global commercial availability of ATOPlex SARS-CoV-2/MPXV/DENV Whole-Genome Targeted Sequencing package compatible with CycloneSEQ sequencing platform.
Microbial Isolates Whole-Genome Sequencing Package: Unlocking the Power of CycloneSEQ
Long-read sequencing technology can complete more complete and continuous genome sequencing when dealing with genomes with large structural variations, high-level repetitive regions, or high GC content. The integration of long-read sequencing technology with short-read sequencing technology can provide a more comprehensive dissection of microbial genomes, offering new insights into the evolution, pathogenicity, and antimicrobial resistance of microorganisms.
Built upon the CycloneSEQ sequencing platform, the package is specifically tailored for microorganisms that have been isolated and cultured. It seamlessly integrates every step of the process, including nucleic acid extraction, library preparation, nanopore sequencing and bioinformatics analysis. This solution not only ensures accurate and comprehensive long-read sequencing of microbial samples, but also automates the analysis of whole-genome assembly, typing, and annotation, delivering a complete end-to-end solution to users.

Figure 1. Microbial Isolates Whole-Genome Sequencing Package (for CycloneSEQ)
The package offers a host of advantages and features that stands out in every step of sequencing workflow to ensure a streamlined sequencing experience:
l Ultra-Fast Library Preparation: The library preparation kit in this package can prepare a single sample library in as little as 40 minutes through a simplified three-step process (end repair →rapid connection of barcode and motor protein → purification). It’s compatible with MGISP-100RS/CyclonePrep-4 for automation system, supporting 1 to 24 samples per run.
l Flexible Testing: Powered by CycloneSEQ-WT02, which features a dual flow cell mode, this package allows for independent operation simultaneously and real-time sequencing with flexible throughput from bp to Mbp levels, meeting the demand for immediate sequencing upon sample receipt.
l Rapid Data Analysis: The software (MGA) in this package can be used for both short-read and long-read sequencing data analysis, and integrates modules for data quality control, assembly, typing, and annotation, allowing for microbial genome analysis in as little as 10 minutes.
l One-stop sequencing experience: From nucleic acid extraction to library preparation, sequencing, and bioinformatics analysis, this package provides a seamless one-stop experience.
Here is a test data of the application for Microbial Isolates Whole-Genome Sequencing Package:
1. High extraction efficiency and high gene integrity
In this study, E. coli bacterial suspension was selected, and the gDNA of E. coli was extracted using the MGIEasy Microbiome DNA&RNA Extraction Kit. The results showed that the DNA extracted by this kit were relatively complete (main band at 23 kb, fragment size > 10 kb) (Figure 2), and has high concentration and purity (Table 1).
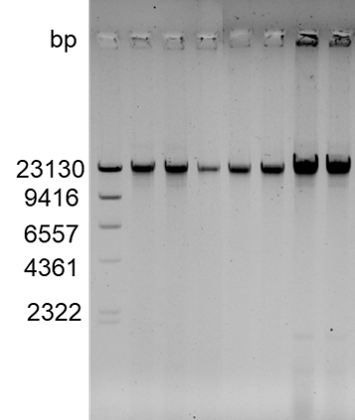
Figure 2. E. coli gDNA bands
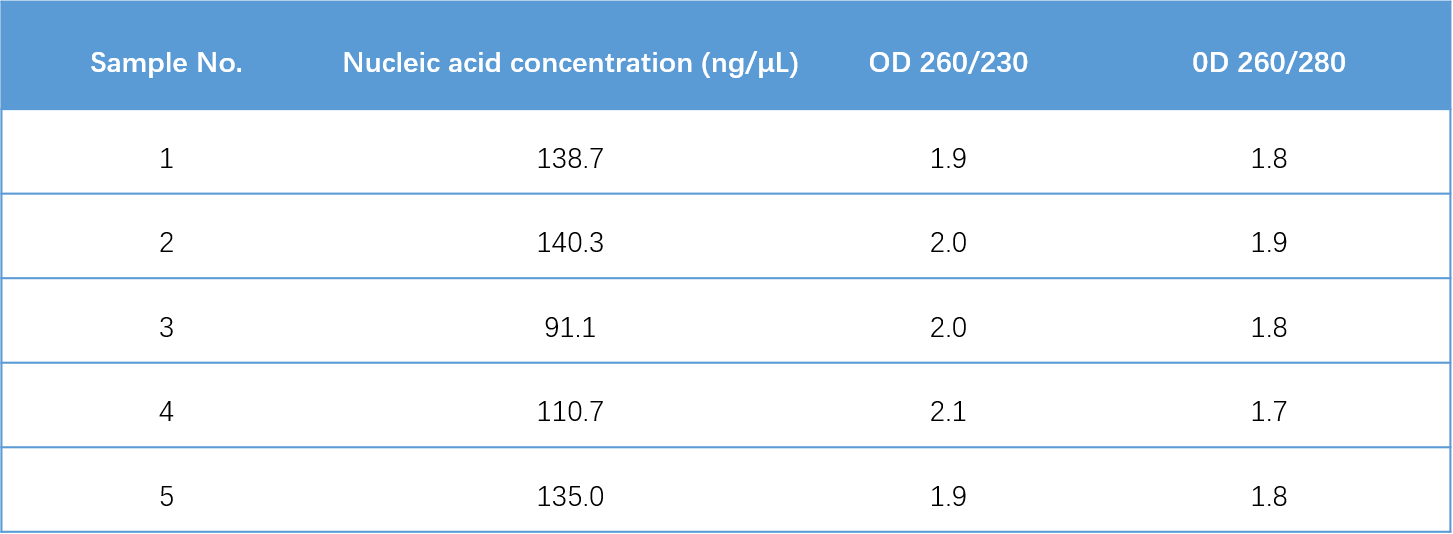
Table 1. Nucleic acid extraction concentration and purity
2. Fewer library preparation steps and higher library yield
Manual methods and the MGISP-100 were used to prepare DNA libraries using the MGIEasy DNA Fast Library Prep Set (for CycloneSEQ).
For each sample, 500 ng of gDNA was input. The experimental results demonstrated that both library preparation methods achieved good efficiency: the yield for manual solution was (52.1 ± 6.8)%, and for automated solution, it was (50.8 ± 3.8)%. Notably, the coefficient of variation (CV) for the automated process (7.4%) was significantly lower than that for manual operation (13.0%), indicating that the automated workflow has better experimental reproducibility (Figure 3).
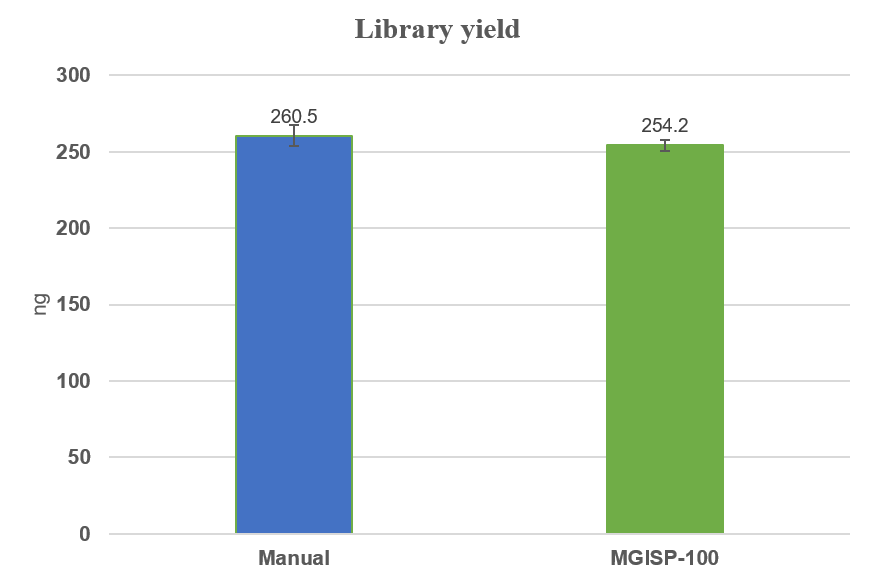
Figure 3. Library yield of manual and automated library preparation
3.Outstanding sequencing quality performance
The 16 E. coli libraries sequenced on the CycloneSEQ-WT02 platform for 48 h generated a total of 22.45 Gb of data, passed N50 is 16.64 Kb, mean length is 5.44 Kb, and Q7 >10 (Table 2). Additionally, the coefficient of variation (CV) for the sequencing data volume was only 8.1%, indicating minimal deviation in the sequencing data output (Figure 4).
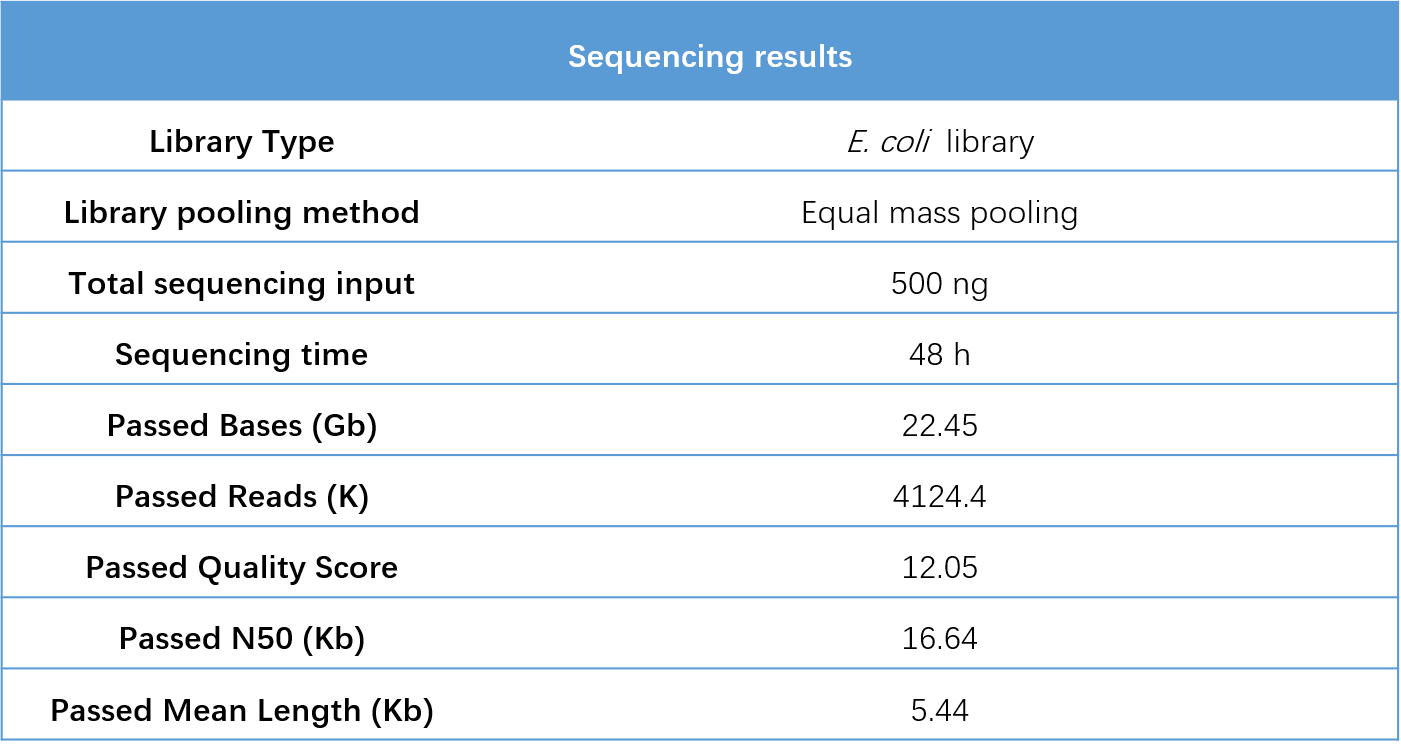
Table 2. Sequencing performance
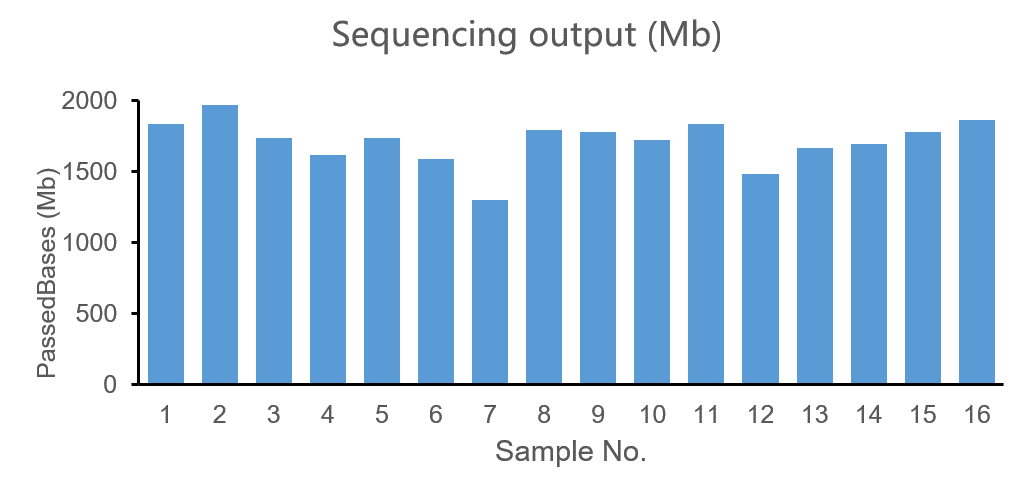
Figure 4. Distribution of sequencing data volume for each sample of the E. coli libraries
4.Excellent whole genome assembly performance
After sequencing on the CycloneSEQ-WT02, the MGA software can automatically initiate analyses such as data quality control, assembly, typing, and annotation.
The analysis results show that the mean length of these long reads is approximately 7 Kb, and the N50 of these long reads are between 15 and 18 Kb. All reads were successfully assembled into circles. The Contig N50 value closely aligns with the total assembly length, and there is just 1 Contig. The lineage identification information is consistent with the known data, and the assembly completeness exceeds 99% (Table 2). All these data demonstrate that this sequencing package can generate low-contamination, high-quality whole-genome data.

Table 3. Sequencing parameters and assembly performance
5.Visualization of Assembly and Functional Prediction
The MGA software can visualize the assembled genomic information to create Circos plots of the assembled sequences, thereby intuitively displaying the structure and relationships of genes (Figure 5). Additionally, the MGA software can perform COG functional gene annotation to predict the potential functions and structures of new gene sequences. This helps us better understand the role of genes in biological processes and provides important references for subsequent gene function research (Figure 5).
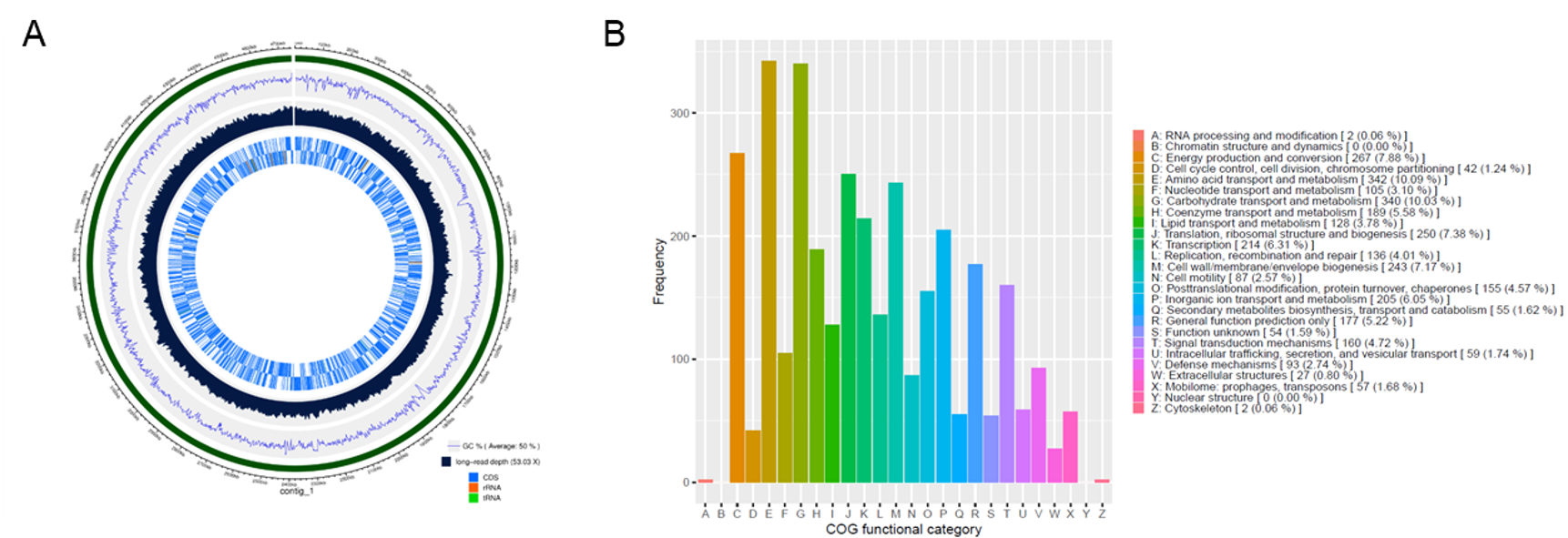
Figure 5. Circos plot and COG functional gene annotation map of E. coli. A. Circos plot have multiple tracks from the outside to the inside. The outermost track represents each assembled contig and its coordinates. The next track is a blue line graph indicating the GC content on the assembled genome. Following that is a black bar graph showing the long-read depth distribution on the assembled genome. The innermost track consists of two concentric highlight bands, indicating the positions of annotated genes on the assembled sequence. Different colors represent distinct genetic elements, with the outer band corresponding to the forward strand and the inner band to the reverse strand. B. The x-axis sequentially arranges the 26 COG clusters, while the y-axis represents the frequency of annotations to different clusters for the sample.
6.Competitive product comparison
Fungal samples and bacterial samples were used to prepare libraries using MGIEasy DNA Fast Library Prep Set (for CycloneSEQ) and library preparation reagents from Brand O, respectively. And then these libraries were sequenced on the CycloneSEQ-WT02 platform and Brand O platform, respectively.
MGI kit only requires 3 steps to prepare the library, which takes 40 minutes, while the O brand kit requires 6 steps to prepare the library, which takes 2 hours and 20 minutes.
After sequencing, the data corresponding to 50× sequencing depth was downsampled for assembly analysis. It was found that for both bacterial and fungal samples, the mean length and N50 of CycloneSEQ-WT02 sequencing sequences were higher than those of brand O , and the final genome assembly was more complete. The above results show that MGIEasy DNA Fast Library Prep Set (for CycloneSEQ) combined with CycloneSEQ platform have high performance.
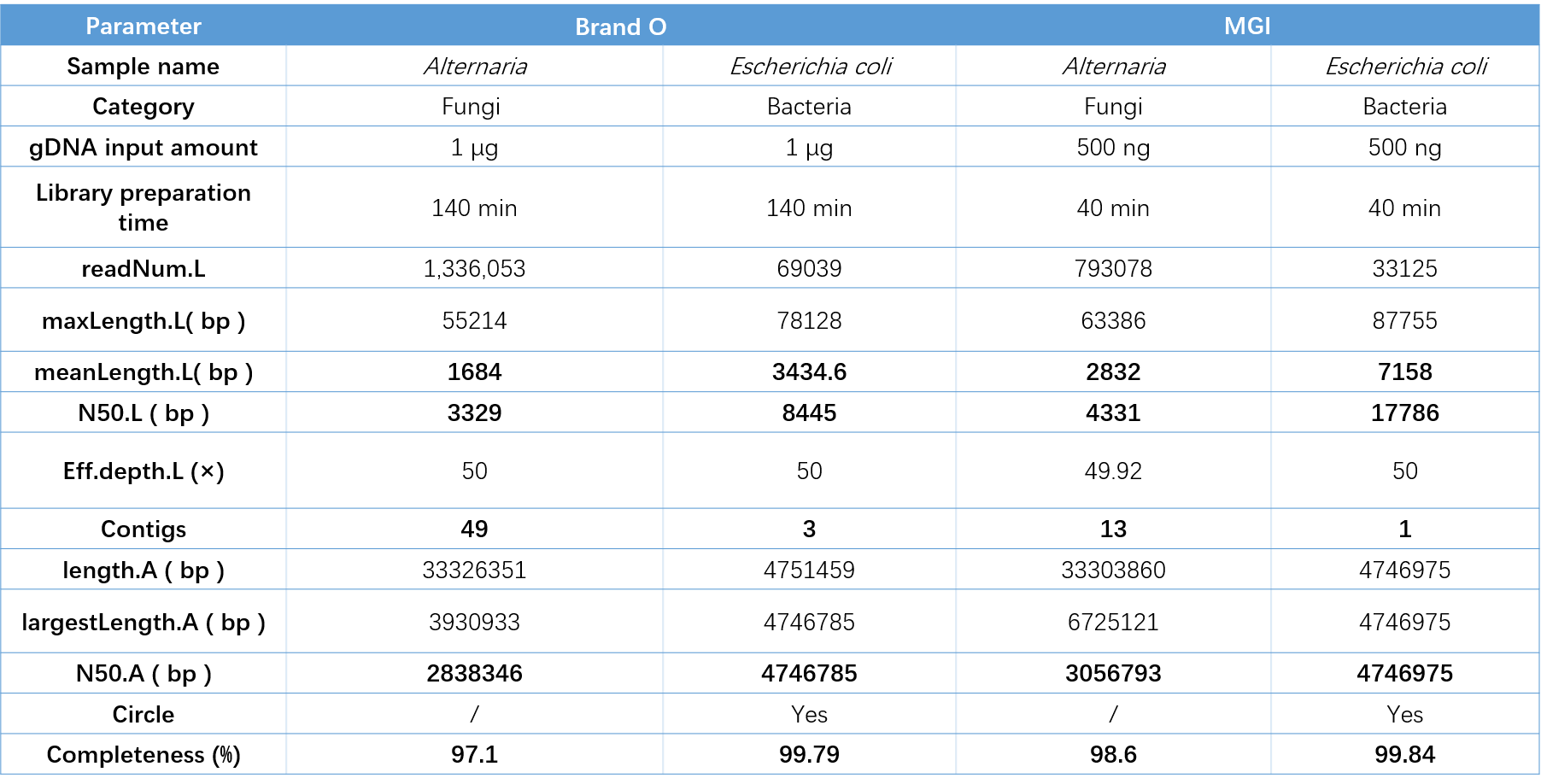
Table 4. Performance comparison between MGI and O brands
ATOPlex SARS-CoV-2/MPXV/DENV Whole Genome Targeted Sequencing Package (for CycloneSEQ): Designed for Pathogen Detection
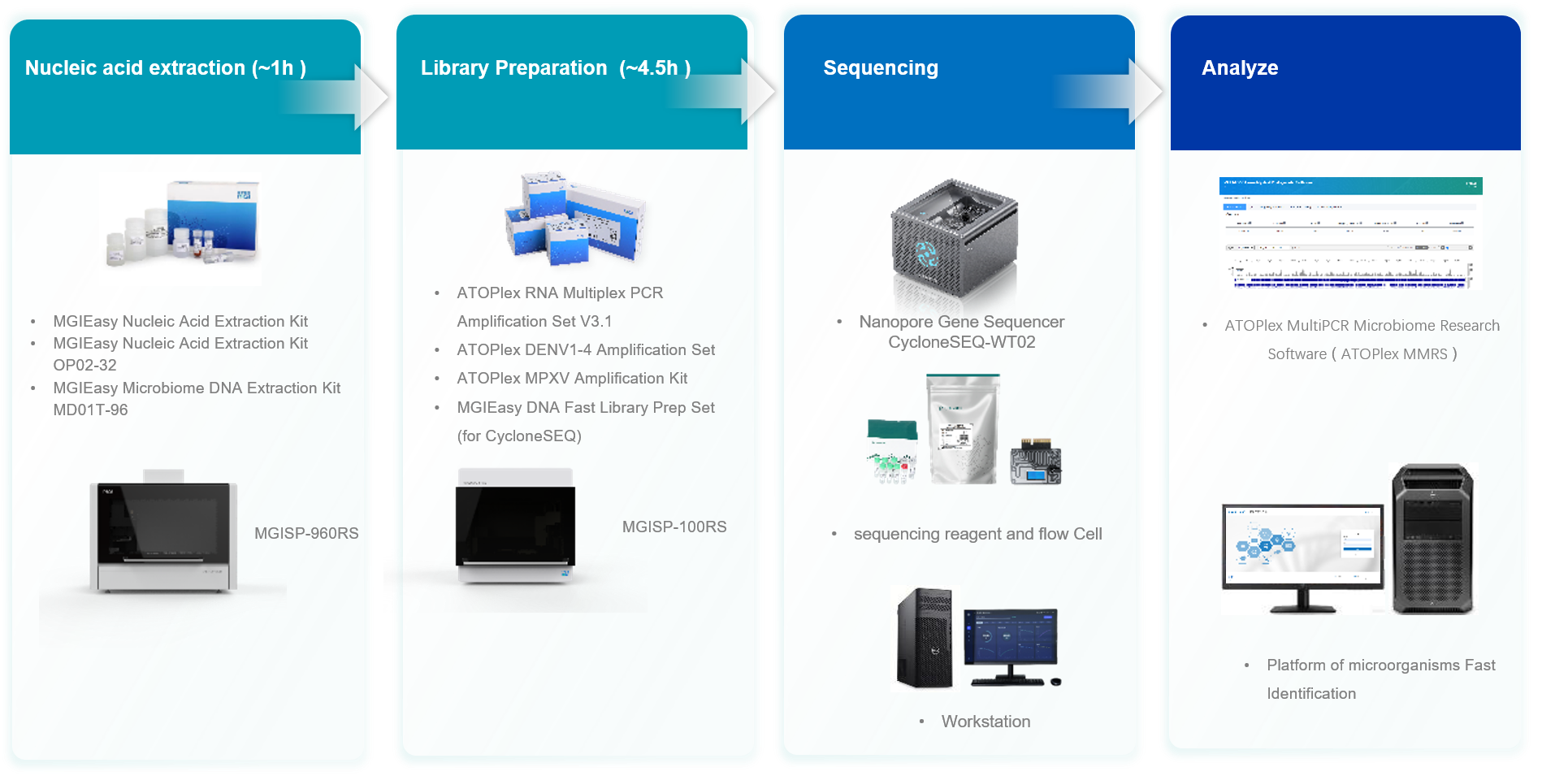
The ATOPlex SARS-CoV-2/MPXV/DENV Whole-Genome Targeted Sequencing Package was created to meet the growing demand for rapid pathogen sequencing, utilizing the CycloneSEQ platform. This sequencing package includes a library preparation kit developed based on the ATOPlex platform (https://en.mgi-tech.com/products/atoplex/). It offers a simple operation and low input requirements, and enables targeted amplification of up to 20,000 target regions in a single tube. Additionally, the panel can be customized to fit specific needs.
The accompanying ATOPlex MMRS software has also been upgraded to fully support the CycloneSEQ-WT02 nanopore sequencer. Benefiting from the CycloneSEQ-WT02's strengths of ultra-long read lengths, real-time sequencing, and the ability to start sequencing without pooling samples, these sequencing packages are ideal for customers handling a small number of samples.
Long-read sequencing, such as CycloneSEQ technology, provides more complete and continuous genome assembly when dealing with genomes that have large structural variations, high levels of repetitive regions, or high GC content. By combining long-read sequencing data with short-read sequencing data, the overall accuracy and completeness of the sequencing data can be further enhanced, providing a more solid data foundation for the classification, functional studies, and evolutionary analysis of microorganisms.
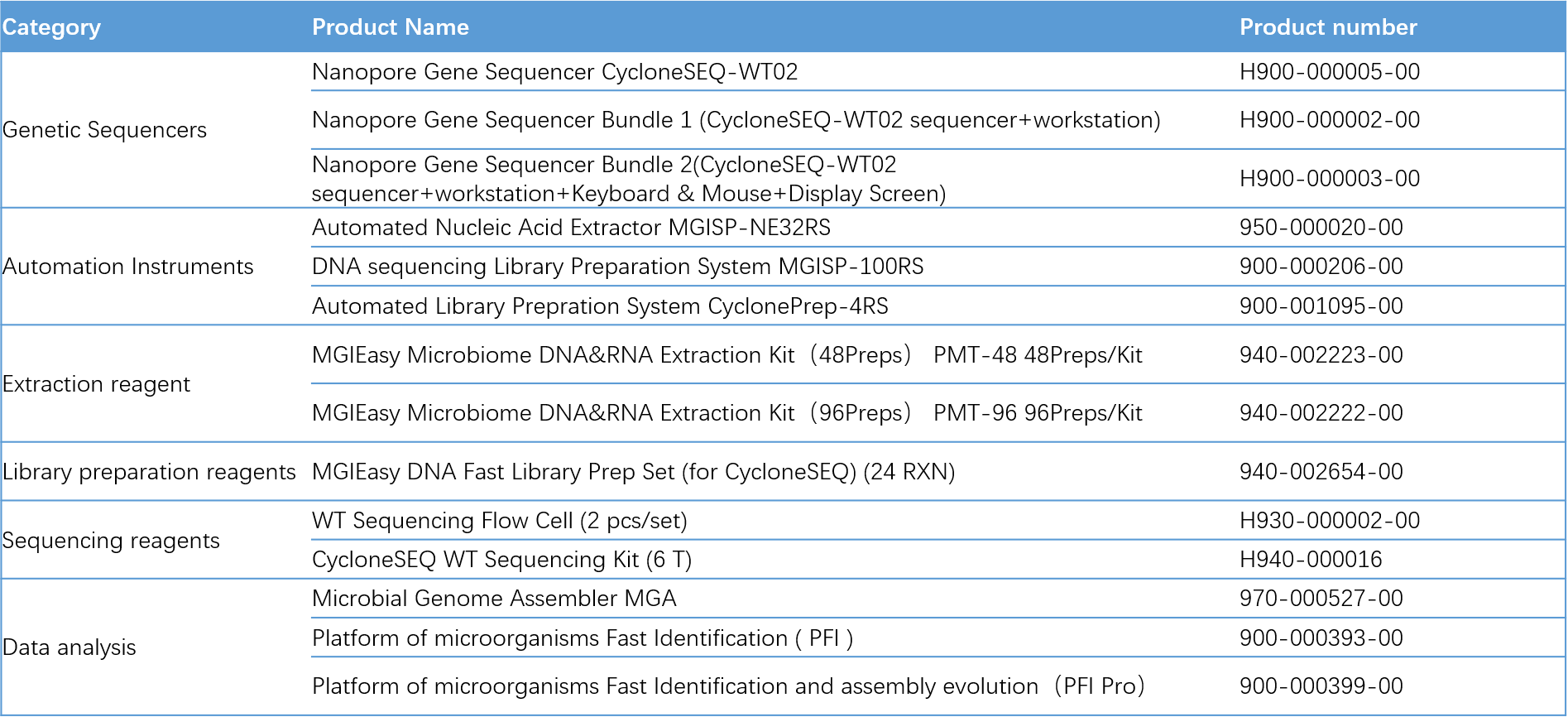
Ordering Information for Microbial Isolates Whole-Genome Sequencing
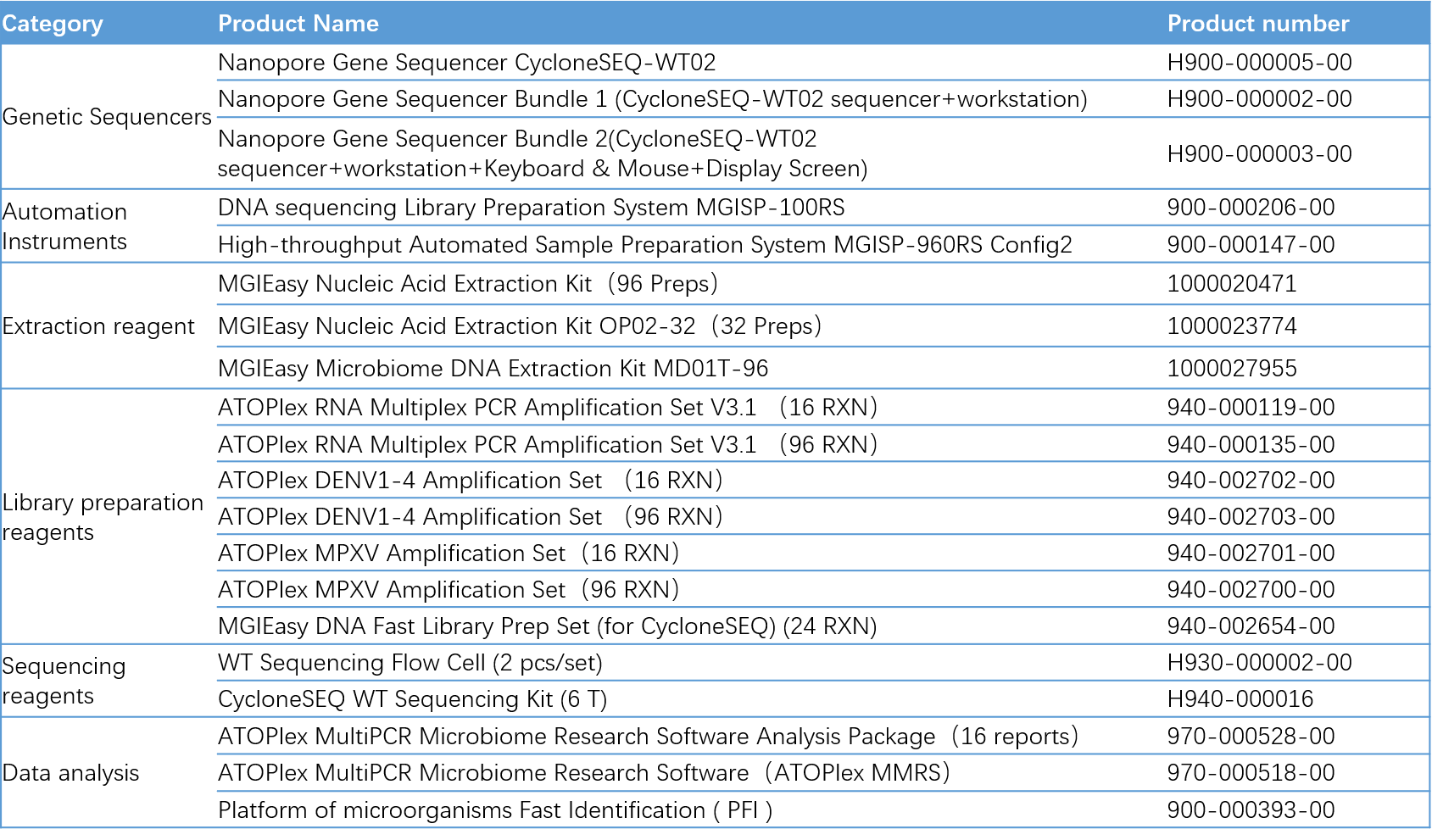
Ordering Information for SARS-CoV-2/MPXV/DENV Whole Genome Targeted Sequencing
Free trial product:
ATOPlex RNA Multiplex PCR Amplification Set V3.1 (16 RXN)
ATOPlex MPXV Amplification Set(16 RXN)
ATOPlex DENV1-4 Amplification Set (16 RXN)
MGIEasy DNA Fast Library Prep Set (for CycloneSEQ) (24 RXN)
Microbial Genome Assembler(MGA) (three-month use right)
ATOPlex MMRS (three-month use right)
Trial application conditions:
Free trial for the first 20 customers (each unit can only apply once)
Trial application date: From now until July 21, 2025
Information registration QR code:

References:
1.Chen L, Zhao N, Cao J, et al. Short-and long-read metagenomics expand individualized structural variations in gut microbiomes[J]. Nature communications, 2022, 13(1): 3175.
2. Liang H, Zou Y, Wang M, et al. Efficiently constructing complete genomes with CycloneSEQ to fill gaps in bacterial draft assemblies[J]. Gigabyte, 2025, 2025: 0-0



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













