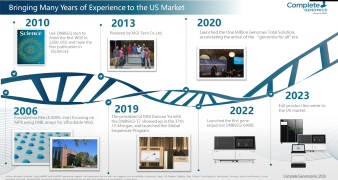
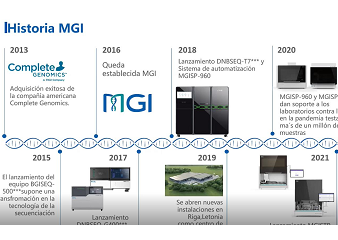
HotMPS is built upon the combinatorial Probe Anchor Synthesis (cPAS) technology found in MGI's proprietary DNBSEQ™ technology. HotMPS inherits the advantages of DNBSEQ™ technology, including low error rate, low duplication rate, and low index hopping, while achieving fundamental breakthroughs in the nucleotides and enzymes used in the sequencing process.
The HotMPS chemistry has significant advantages of stronger signal, less systematic sequence-based errors and could be compatible with commonly used library preparation methods.
Improving the quality while lowering the cost of high-throughput sequencing through these new technologies can help to drive the implementation of genomics-based health monitoring and other applications that require comprehensive, accurate and affordable sequencing-based tests. MGI is continuously advancing MPS technology to empower our partners, customers and the industry to work toward a full understanding of the human and other species genomes and cells, and to ultimately improve our health.
* This sequencer is only available in selected countries, and its software has been specially configured to be used in conjunction with MGI’s HotMPS sequencing reagents exclusively.
**This sequencing reagent is only available in selected countries.
If you have any questions, you can send an email to:



 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL















 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club